The structure of human GALNS reveals the molecular basis for mucopolysaccharidosis IV A
- PMID: 22940367
- PMCID: PMC3472114
- DOI: 10.1016/j.jmb.2012.08.020
The structure of human GALNS reveals the molecular basis for mucopolysaccharidosis IV A
Abstract
Lysosomal enzymes catalyze the breakdown of macromolecules in the cell. In humans, loss of activity of a lysosomal enzyme leads to an inherited metabolic defect known as a lysosomal storage disorder. The human lysosomal enzyme galactosamine-6-sulfatase (GALNS, also known as N-acetylgalactosamine-6-sulfatase and GalN6S; E.C. 3.1.6.4) is deficient in patients with the lysosomal storage disease mucopolysaccharidosis IV A (also known as MPS IV A and Morquio A). Here, we report the three-dimensional structure of human GALNS, determined by X-ray crystallography at 2.2Å resolution. The structure reveals a catalytic gem diol nucleophile derived from modification of a cysteine side chain. The active site of GALNS is a large, positively charged trench suitable for binding polyanionic substrates such as keratan sulfate and chondroitin-6-sulfate. Enzymatic assays on the insect-cell-expressed human GALNS indicate activity against synthetic substrates and inhibition by both substrate and product. Mapping 120 MPS IV A missense mutations onto the structure reveals that a majority of mutations affect the hydrophobic core of the structure, indicating that most MPS IV A cases result from misfolding of GALNS. Comparison of the structure of GALNS to paralogous sulfatases shows a wide variety of active-site geometries in the family but strict conservation of the catalytic machinery. Overall, the structure and the known mutations establish the molecular basis for MPS IV A and for the larger MPS family of diseases.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures





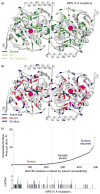
References
-
- Neufeld EF, Muenzer J. The Mucopolysaccharidoses. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. 8. McGraw-Hill; New York: 2001. pp. 3421–3452.
-
- Northover H, Cowie RA, Wraith JE. Mucopolysaccharidosis type IVA (Morquio syndrome): a clinical review. J Inherit Metab Dis. 1996;19:357–65. - PubMed
-
- Tomatsu S, Montano AM, Nishioka T, Gutierrez MA, Pena OM, Tranda Firescu GG, Lopez P, Yamaguchi S, Noguchi A, Orii T. Mutation and polymorphism spectrum of the GALNS gene in mucopolysaccharidosis IVA (Morquio A) Hum Mutat. 2005;26:500–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

