Electrical activity can impose time of day on the circadian transcriptome of pacemaker neurons
- PMID: 22940468
- PMCID: PMC3562355
- DOI: 10.1016/j.cub.2012.07.070
Electrical activity can impose time of day on the circadian transcriptome of pacemaker neurons
Abstract
Background: Circadian (∼24 hr) rhythms offer one of the best examples of how gene expression is tied to behavior. Circadian pacemaker neurons contain molecular clocks that control 24 hr rhythms in gene expression that in turn regulate electrical activity rhythms to control behavior.
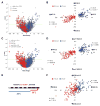
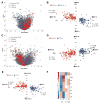
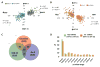
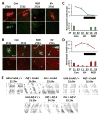
Results: Here we demonstrate the inverse relationship: there are broad transcriptional changes in Drosophila clock neurons (LN(v)s) in response to altered electrical activity, including a large set of circadian genes. Hyperexciting LN(v)s creates a morning-like expression profile for many circadian genes while hyperpolarization leads to an evening-like transcriptional state. The electrical effects robustly persist in per(0) mutant LN(v)s but not in cyc(0) mutant LN(v)s, suggesting that neuronal activity interacts with the transcriptional activators of the core circadian clock. Bioinformatic and immunocytochemical analyses suggest that CREB family transcription factors link LN(v) electrical state to circadian gene expression.
Conclusions: The electrical state of a clock neuron can impose time of day to its transcriptional program. We propose that this acts as an internal zeitgeber to add robustness and precision to circadian behavioral rhythms.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures

Comment in
-
Circadian rhythms: An electric jolt to the clock.Curr Biol. 2012 Oct 23;22(20):R876-8. doi: 10.1016/j.cub.2012.08.003. Curr Biol. 2012. PMID: 23098596
References
-
- Guan Z, Saraswati S, Adolfsen B, Littleton JT. Genome-wide transcriptional changes associated with enhanced activity in the Drosophila nervous system. Neuron. 2005;48:91–107. - PubMed
-
- Deisseroth K, Bito H, Tsien RW. Signaling from synapse to nucleus: postsynaptic CREB phosphorylation during multiple forms of hippocampal synaptic plasticity. Neuron. 1996;16:89–101. - PubMed
-
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

