OmpA-mediated biofilm formation is essential for the commensal bacterium Sodalis glossinidius to colonize the tsetse fly gut
- PMID: 22941073
- PMCID: PMC3485708
- DOI: 10.1128/AEM.01858-12
OmpA-mediated biofilm formation is essential for the commensal bacterium Sodalis glossinidius to colonize the tsetse fly gut
Abstract
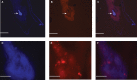
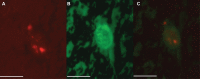
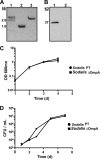
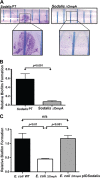
Many bacteria successfully colonize animals by forming protective biofilms. Molecular processes that underlie the formation and function of biofilms in pathogenic bacteria are well characterized. In contrast, the relationship between biofilms and host colonization by symbiotic bacteria is less well understood. Tsetse flies (Glossina spp.) house 3 maternally transmitted symbionts, one of which is a commensal (Sodalis glossinidius) found in several host tissues, including the gut. We determined that Sodalis forms biofilms in the tsetse gut and that this process is influenced by the Sodalis outer membrane protein A (OmpA). Mutant Sodalis strains that do not produce OmpA (Sodalis ΔOmpA mutants) fail to form biofilms in vitro and are unable to colonize the tsetse gut unless endogenous symbiotic bacteria are present. Our data indicate that in the absence of biofilms, Sodalis ΔOmpA mutant cells are exposed to and eliminated by tsetse's innate immune system, suggesting that biofilms help Sodalis evade the host immune system. Tsetse is the sole vector of pathogenic African trypanosomes, which also reside in the fly gut. Acquiring a better understanding of the dynamics that promote Sodalis colonization of the tsetse gut may enhance the development of novel disease control strategies.
Figures

References
-
- Akman L, et al. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32:402–407 - PubMed
-
- Aksoy S, Pourhosseini AA, Chow A. 1995. Mycetome endosymbionts of tsetse flies constitute a distinct lineage related to Enterobacteriaceae. Insect Mol. Biol. 4:15–22 - PubMed
-
- Barrios AF, Zuo R, Ren D, Wood TK. 2006. Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol. Bioeng. 93:188–200 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

