Measuring the formation and repair of UV damage at the DNA sequence level by ligation-mediated PCR
- PMID: 22941605
- PMCID: PMC3543819
- DOI: 10.1007/978-1-61779-998-3_14
Measuring the formation and repair of UV damage at the DNA sequence level by ligation-mediated PCR
Abstract
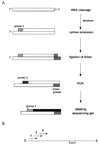
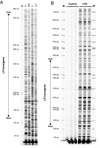
The formation and repair of DNA damage at specific locations in the genome is modulated by DNA sequence context, by DNA cytosine-5 methylation patterns, by the transcriptional status of the locus and by proteins associated with the DNA. The only method currently available to allow precise sequence mapping of DNA lesions in mammalian cells is the ligation-mediated polymerase chain reaction (LM-PCR) technique. We provide an update on technical details of LM-PCR. LM-PCR can be used, for example, for mapping of ultraviolet (UV) light-induced DNA photoproducts such as cyclobutane pyrimidine dimers.
Figures
Similar articles
-
Measuring the formation and repair of DNA damage by ligation-mediated PCR.Methods Mol Biol. 2006;314:201-14. doi: 10.1385/1-59259-973-7:201. Methods Mol Biol. 2006. PMID: 16673883
-
Measuring the formation and repair of UV photoproducts by ligation-mediated PCR.Methods Mol Biol. 1999;113:213-26. doi: 10.1385/1-59259-675-4:213. Methods Mol Biol. 1999. PMID: 10443423 No abstract available.
-
Measurement of activities of cyclobutane-pyrimidine-dimer and (6-4)-photoproduct photolyases.Methods Mol Biol. 1999;113:133-46. doi: 10.1385/1-59259-675-4:133. Methods Mol Biol. 1999. PMID: 10443416 No abstract available.
-
Repair of (6-4) Lesions in DNA by (6-4) Photolyase: 20 Years of Quest for the Photoreaction Mechanism.Photochem Photobiol. 2017 Jan;93(1):51-66. doi: 10.1111/php.12696. Epub 2017 Jan 31. Photochem Photobiol. 2017. PMID: 27992654 Review.
-
Detection of DNA adducts at the DNA sequence level by ligation-mediated PCR.Mutat Res. 1993 Jul;288(1):39-46. doi: 10.1016/0027-5107(93)90206-u. Mutat Res. 1993. PMID: 7686264 Review.
Cited by
-
Rates of chemical cleavage of DNA and RNA oligomers containing guanine oxidation products.Chem Res Toxicol. 2015 Jun 15;28(6):1292-300. doi: 10.1021/acs.chemrestox.5b00096. Epub 2015 Apr 22. Chem Res Toxicol. 2015. PMID: 25853314 Free PMC article.
-
Molecular mechanisms and genomic maps of DNA excision repair in Escherichia coli and humans.J Biol Chem. 2017 Sep 22;292(38):15588-15597. doi: 10.1074/jbc.R117.807453. Epub 2017 Aug 10. J Biol Chem. 2017. PMID: 28798238 Free PMC article. Review.
-
Quantifying DNA double-strand breaks induced by site-specific endonucleases in living cells by ligation-mediated purification.Nat Protoc. 2014 Mar;9(3):517-28. doi: 10.1038/nprot.2014.031. Epub 2014 Feb 6. Nat Protoc. 2014. PMID: 24504477
-
Dynamic maps of UV damage formation and repair for the human genome.Proc Natl Acad Sci U S A. 2017 Jun 27;114(26):6758-6763. doi: 10.1073/pnas.1706522114. Epub 2017 Jun 12. Proc Natl Acad Sci U S A. 2017. PMID: 28607063 Free PMC article.
-
Detecting Rare Mutations and DNA Damage with Sequencing-Based Methods.Trends Biotechnol. 2018 Jul;36(7):729-740. doi: 10.1016/j.tibtech.2018.02.009. Epub 2018 Mar 14. Trends Biotechnol. 2018. PMID: 29550161 Free PMC article. Review.
References
-
- Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571:19–31. - PubMed
-
- Yoon J-H, Lee C-S, O’Connor T, Yasui A, Pfeifer GP. The DNA damage spectrum produced by simulated sunlight. J Mol Biol. 2000;299:681–693. - PubMed
-
- You YH, Lee DH, Yoon JH, Nakajima S, Yasui A, Pfeifer GP. Cyclobutane pyrimidine dimers are responsible for the vast majority of mutations induced by UVB irradiation in mammalian cells. J Biol Chem. 2001;276:44688–44694. - PubMed
-
- Mitchell DL, Nairn RS. The biology of the (6-4) photoproduct. Photochem Photobiol. 1989;49:805–819. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

