Mitochondrial DNA copy number is regulated in a tissue specific manner by DNA methylation of the nuclear-encoded DNA polymerase gamma A
- PMID: 22941637
- PMCID: PMC3488228
- DOI: 10.1093/nar/gks770
Mitochondrial DNA copy number is regulated in a tissue specific manner by DNA methylation of the nuclear-encoded DNA polymerase gamma A
Abstract
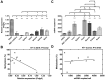
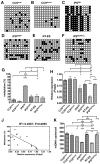
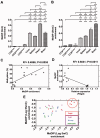
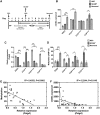
DNA methylation is an essential mechanism controlling gene expression during differentiation and development. We investigated the epigenetic regulation of the nuclear-encoded, mitochondrial DNA (mtDNA) polymerase γ catalytic subunit (PolgA) by examining the methylation status of a CpG island within exon 2 of PolgA. Bisulphite sequencing identified low methylation levels (<10%) within exon 2 of mouse oocytes, blastocysts and embryonic stem cells (ESCs), while somatic tissues contained significantly higher levels (>40%). In contrast, induced pluripotent stem (iPS) cells and somatic nuclear transfer ESCs were hypermethylated (>20%), indicating abnormal epigenetic reprogramming. Real time PCR analysis of 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) immunoprecipitated DNA suggests active DNA methylation and demethylation within exon 2 of PolgA. Moreover, neural differentiation of ESCs promoted de novo methylation and demethylation at the exon 2 locus. Regression analysis demonstrates that cell-specific PolgA expression levels were negatively correlated with DNA methylation within exon 2 and mtDNA copy number. Finally, using chromatin immunoprecipitation (ChIP) against RNA polymerase II (RNApII) phosphorylated on serine 2, we show increased DNA methylation levels are associated with reduced RNApII transcriptional elongation. This is the first study linking nuclear DNA epigenetic regulation with mtDNA regulation during differentiation and cell specialization.
Figures


References
-
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
-
- Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. - PubMed
-
- Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim. Biophys. Acta. 2010;1797:113–128. - PubMed
-
- Ropp PA, Copeland WC. Cloning and characterization of the human mitochondrial DNA polymerase, DNA polymerase gamma. Genomics. 1996;36:449–458. - PubMed
-
- Sweasy JB, Lauper JM, Eckert KA. DNA polymerases and human diseases. Radiat. Res. 2006;166:693–714. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

