PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks
- PMID: 22941645
- PMCID: PMC3488241
- DOI: 10.1093/nar/gks798
PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks
Abstract
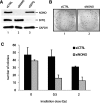
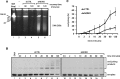
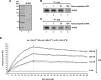
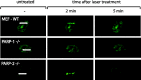
After the generation of DNA double-strand breaks (DSBs), poly(ADP-ribose) polymerase-1 (PARP-1) is one of the first proteins to be recruited and activated through its binding to the free DNA ends. Upon activation, PARP-1 uses NAD+ to generate large amounts of poly(ADP-ribose) (PAR), which facilitates the recruitment of DNA repair factors. Here, we identify the RNA-binding protein NONO, a partner protein of SFPQ, as a novel PAR-binding protein. The protein motif being primarily responsible for PAR-binding is the RNA recognition motif 1 (RRM1), which is also crucial for RNA-binding, highlighting a competition between RNA and PAR as they share the same binding site. Strikingly, the in vivo recruitment of NONO to DNA damage sites completely depends on PAR, generated by activated PARP-1. Furthermore, we show that upon PAR-dependent recruitment, NONO stimulates nonhomologous end joining (NHEJ) and represses homologous recombination (HR) in vivo. Our results therefore place NONO after PARP activation in the context of DNA DSB repair pathway decision. Understanding the mechanism of action of proteins that act in the same pathway as PARP-1 is crucial to shed more light onto the effect of interference on PAR-mediated pathways with PARP inhibitors, which have already reached phase III clinical trials but are until date poorly understood.
Figures



Similar articles
-
IGFBP-3 interacts with NONO and SFPQ in PARP-dependent DNA damage repair in triple-negative breast cancer.Cell Mol Life Sci. 2019 May;76(10):2015-2030. doi: 10.1007/s00018-019-03033-4. Epub 2019 Feb 6. Cell Mol Life Sci. 2019. PMID: 30725116 Free PMC article.
-
SFPQ•NONO and XLF function separately and together to promote DNA double-strand break repair via canonical nonhomologous end joining.Nucleic Acids Res. 2017 Feb 28;45(4):1848-1859. doi: 10.1093/nar/gkw1209. Nucleic Acids Res. 2017. PMID: 27924002 Free PMC article.
-
Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response.Cell Cycle. 2010 Apr 15;9(8):1568-76. doi: 10.4161/cc.9.8.11298. Epub 2010 Apr 15. Cell Cycle. 2010. PMID: 20421735
-
Multiple functions of PARP1 in the repair of DNA double strand breaks.DNA Repair (Amst). 2025 Aug;152:103873. doi: 10.1016/j.dnarep.2025.103873. Epub 2025 Jul 21. DNA Repair (Amst). 2025. PMID: 40712289 Review.
-
Structure and function of poly(ADP-ribose) polymerase-1: role in oxidative stress-related pathologies.Curr Vasc Pharmacol. 2005 Jul;3(3):209-14. doi: 10.2174/1570161054368625. Curr Vasc Pharmacol. 2005. PMID: 16026317 Review.
Cited by
-
OGA is associated with deglycosylation of NONO and the KU complex during DNA damage repair.Cell Death Dis. 2021 Jun 16;12(7):622. doi: 10.1038/s41419-021-03910-6. Cell Death Dis. 2021. PMID: 34135314 Free PMC article.
-
Break-induced RNA-DNA hybrids (BIRDHs) in homologous recombination: friend or foe?EMBO Rep. 2023 Dec 6;24(12):e57801. doi: 10.15252/embr.202357801. Epub 2023 Oct 11. EMBO Rep. 2023. PMID: 37818834 Free PMC article. Review.
-
Maintenance of genome stability: the unifying role of interconnections between the DNA damage response and RNA-processing pathways.Curr Genet. 2018 Oct;64(5):971-983. doi: 10.1007/s00294-018-0819-7. Epub 2018 Mar 1. Curr Genet. 2018. PMID: 29497809 Review.
-
Why structure and chain length matter: on the biological significance underlying the structural heterogeneity of poly(ADP-ribose).Nucleic Acids Res. 2021 Sep 7;49(15):8432-8448. doi: 10.1093/nar/gkab618. Nucleic Acids Res. 2021. PMID: 34302489 Free PMC article.
-
USP39 promotes non-homologous end-joining repair by poly(ADP-ribose)-induced liquid demixing.Nucleic Acids Res. 2021 Nov 8;49(19):11083-11102. doi: 10.1093/nar/gkab892. Nucleic Acids Res. 2021. PMID: 34614178 Free PMC article.
References
-
- Wacker DA, Frizzell KM, Zhang T, Kraus WL. Regulation of chromatin structure and chromatin-dependent transcription by poly(ADP-ribose) polymerase-1: possible targets for drug-based therapies. Subcell Biochem. 2007;41:45–69. - PubMed
-
- Bouchard VJ, Rouleau M, Poirier GG. PARP-1, a determinant of cell survival in response to DNA damage. Exp. Hematol. 2003;31:446–454. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

