DOT1A-dependent H3K76 methylation is required for replication regulation in Trypanosoma brucei
- PMID: 22941659
- PMCID: PMC3488242
- DOI: 10.1093/nar/gks801
DOT1A-dependent H3K76 methylation is required for replication regulation in Trypanosoma brucei
Abstract
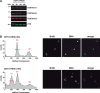
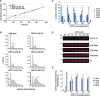
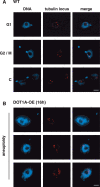
Cell-cycle progression requires careful regulation to ensure accurate propagation of genetic material to the daughter cells. Although many cell-cycle regulators are evolutionarily conserved in the protozoan parasite Trypanosoma brucei, novel regulatory mechanisms seem to have evolved. Here, we analyse the function of the histone methyltransferase DOT1A during cell-cycle progression. Over-expression of DOT1A generates a population of cells with aneuploid nuclei as well as enucleated cells. Detailed analysis shows that DOT1A over-expression causes continuous replication of the nuclear DNA. In contrast, depletion of DOT1A by RNAi abolishes replication but does not prevent karyokinesis. As histone H3K76 methylation has never been associated with replication control in eukaryotes before, we have discovered a novel function of DOT1 enzymes, which might not be unique to trypanosomes.
Figures



Similar articles
-
Heterologous expression reveals distinct enzymatic activities of two DOT1 histone methyltransferases of Trypanosoma brucei.J Cell Sci. 2010 Dec 1;123(Pt 23):4019-23. doi: 10.1242/jcs.073882. J Cell Sci. 2010. PMID: 21084562
-
Dot1 histone methyltransferases share a distributive mechanism but have highly diverged catalytic properties.Sci Rep. 2015 May 12;5:9824. doi: 10.1038/srep09824. Sci Rep. 2015. PMID: 25965993 Free PMC article.
-
Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei.Mol Cell. 2006 Aug;23(4):497-507. doi: 10.1016/j.molcel.2006.06.027. Mol Cell. 2006. PMID: 16916638
-
Regulation of the cell division cycle in Trypanosoma brucei.Eukaryot Cell. 2012 Oct;11(10):1180-90. doi: 10.1128/EC.00145-12. Epub 2012 Aug 3. Eukaryot Cell. 2012. PMID: 22865501 Free PMC article. Review.
-
Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity.Nucleic Acids Res. 2013 Mar 1;41(5):2797-806. doi: 10.1093/nar/gkt012. Epub 2013 Jan 23. Nucleic Acids Res. 2013. PMID: 23345616 Free PMC article. Review.
Cited by
-
Methylation of histone H3 on lysine 79 associates with a group of replication origins and helps limit DNA replication once per cell cycle.PLoS Genet. 2013 Jun;9(6):e1003542. doi: 10.1371/journal.pgen.1003542. Epub 2013 Jun 6. PLoS Genet. 2013. PMID: 23754963 Free PMC article.
-
Unexpected diversity in eukaryotic transcription revealed by the retrotransposon hotspot family of Trypanosoma brucei.Nucleic Acids Res. 2019 Feb 28;47(4):1725-1739. doi: 10.1093/nar/gky1255. Nucleic Acids Res. 2019. PMID: 30544263 Free PMC article.
-
TelAP1 links telomere complexes with developmental expression site silencing in African trypanosomes.Nucleic Acids Res. 2018 Apr 6;46(6):2820-2833. doi: 10.1093/nar/gky028. Nucleic Acids Res. 2018. PMID: 29385523 Free PMC article.
-
The many faces of histone H3K79 methylation.Mutat Res Rev Mutat Res. 2016 Apr-Jun;768:46-52. doi: 10.1016/j.mrrev.2016.03.005. Epub 2016 Mar 31. Mutat Res Rev Mutat Res. 2016. PMID: 27234562 Free PMC article. Review.
-
Trypanosomes can initiate nuclear export co-transcriptionally.Nucleic Acids Res. 2019 Jan 10;47(1):266-282. doi: 10.1093/nar/gky1136. Nucleic Acids Res. 2019. PMID: 30418648 Free PMC article.
References
-
- Janzen CJ, Hake SB, Lowell JE, Cross GA. Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol. Cell. 2006;23:497–507. - PubMed
-
- van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. - PubMed
-
- Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 2006;281:559–568. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

