Comprehensive functional analysis of chymotrypsin C (CTRC) variants reveals distinct loss-of-function mechanisms associated with pancreatitis risk
- PMID: 22942235
- PMCID: PMC3660471
- DOI: 10.1136/gutjnl-2012-303090
Comprehensive functional analysis of chymotrypsin C (CTRC) variants reveals distinct loss-of-function mechanisms associated with pancreatitis risk
Abstract
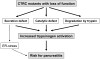
Objective: The digestive enzyme chymotrypsin C (CTRC) protects against pancreatitis by promoting degradation of trypsinogen, thereby curtailing potentially harmful trypsinogen activation. Loss-of-function variants in CTRC increase the risk for chronic pancreatitis. The aim of the present study was to perform comprehensive functional analysis of all missense CTRC variants identified to date.
Design: We investigated secretion, activity and degradation of 27 published and five novel CTRC mutants. We also assessed the effect of five mutants on endoplasmic reticulum (ER) stress.
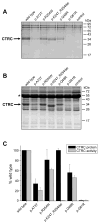
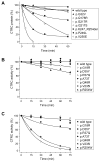
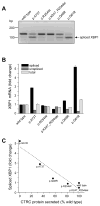
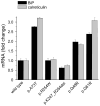
Results: None of the mutants exhibited a gain of function, such as increased secretion or activity. By contrast, 11 mutants showed marked loss of function, three mutants had moderate functional defects, whereas 18 mutants were functionally similar to wild-type CTRC. The functional deficiencies observed were diminished secretion, impaired catalytic activity and degradation by trypsin. Mutants with a secretion defect caused ER stress that was proportional to the loss in secretion. ER stress was not associated with loss-of-function phenotypes related to catalytic defect or proteolytic instability.
Conclusions: Pathogenic CTRC variants cause loss of function by three distinct but mutually non-exclusive mechanisms that affect secretion, activity and proteolytic stability. ER stress may be induced by a subset of CTRC mutants, but does not represent a common pathological mechanism of CTRC variants. This phenotypic dataset should aid in the classification of the clinical relevance of CTRC variants identified in patients with chronic pancreatitis.
Keywords: Pancreatic Disease; Pancreatic Disorders; Pancreatic Enzymes; Pancreatic Physiology; Pancreatitis.
Conflict of interest statement
No competing interest to declare.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
