Ribulose-1,5-bis-phosphate carboxylase/oxygenase accumulation factor1 is required for holoenzyme assembly in maize
- PMID: 22942379
- PMCID: PMC3462642
- DOI: 10.1105/tpc.112.102012
Ribulose-1,5-bis-phosphate carboxylase/oxygenase accumulation factor1 is required for holoenzyme assembly in maize
Abstract
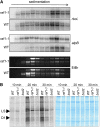
Most life is ultimately sustained by photosynthesis and its rate-limiting carbon fixing enzyme, ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco). Although the structurally comparable cyanobacterial Rubisco is amenable to in vitro assembly, the higher plant enzyme has been refractory to such manipulation due to poor understanding of its assembly pathway. Here, we report the identification of a chloroplast protein required for Rubisco accumulation in maize (Zea mays), RUBISCO ACCUMULATION FACTOR1 (RAF1), which lacks any characterized functional domains. Maize lines lacking RAF1 due to Mutator transposon insertions are Rubisco deficient and seedling lethal. Analysis of transcripts and proteins showed that Rubisco large subunit synthesis in raf1 plants is not compromised; however, newly synthesized Rubisco large subunit appears in a high molecular weight form whose accumulation requires a specific chaperonin 60 isoform. Gel filtration analysis and blue native gels showed that endogenous and recombinant RAF1 are trimeric; however, following in vivo cross-linking, RAF1 copurifies with Rubisco large subunit, suggesting that they interact weakly or transiently. RAF1 is predominantly expressed in bundle sheath chloroplasts, consistent with a Rubisco accumulation function. Our results support the hypothesis that RAF1 acts during Rubisco assembly by releasing and/or sequestering the large subunit from chaperonins early in the assembly process.
Figures





Comment in
-
Insight into ribulose 1,5-bis-phosphate carboxylase/oxygenase assembly in maize.Plant Cell. 2012 Aug;24(8):3171. doi: 10.1105/tpc.112.240814. Epub 2012 Aug 31. Plant Cell. 2012. PMID: 22942384 Free PMC article. No abstract available.
-
The first chaperonin.Nat Rev Mol Cell Biol. 2013 Oct;14(10):611. doi: 10.1038/nrm3665. Nat Rev Mol Cell Biol. 2013. PMID: 24061226 No abstract available.
References
-
- Andersson I., Backlund A. (2008). Structure and function of Rubisco. Plant Physiol. Biochem. 46: 275–291 - PubMed
-
- Andrews T.J. (1988). Catalysis by cyanobacterial ribulose-bisphosphate carboxylase large subunits in the complete absence of small subunits. J. Biol. Chem. 263: 12213–12219 - PubMed
-
- Bannai H., Tamada Y., Maruyama O., Nakai K., Miyano S. (2002). Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18: 298–305 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

