Unfolding analysis of the mature and unprocessed forms of Bacillus licheniformis γ-glutamyltranspeptidase
- PMID: 22942488
- PMCID: PMC3169692
- DOI: 10.1007/s10867-011-9228-6
Unfolding analysis of the mature and unprocessed forms of Bacillus licheniformis γ-glutamyltranspeptidase
Abstract
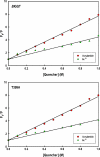
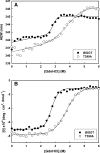
Bacillus licheniformis γ-glutamyltranspeptidase (BlGGT) undergoes an autocatalytic process to generate 44.9 and 21.7 kDa subunits; however, a mutant protein (T399A) loses completely the processing ability and mainly exists as a precursor. For a comprehensive understanding of their structural features, the biophysical properties of these two proteins were investigated by circular dichroism and fluorescence spectroscopy. Tryptophan fluorescence and circular dichroism spectra were nearly identical for BlGGT and T399A, but unfolding analyses revealed that these two proteins had a different sensitivity towards temperature- and guanidine hydrochloride (GdnHCl)-induced denaturation. BlGGT and the unprocessed T399A displayed T(m) values of 61.4°C and 68.1°C, respectively, and thermal unfolding of both proteins was found to be highly irreversible. Fluorescence quenching analysis showed that T399A had a dynamic quenching constant similar to that of the wild-type enzyme. BlGGT started to unfold beyond ∼2.14 M GdnHCl and reached an unfolded intermediate, [GdnHCl](0.5, N - U), at 2.85 M, corresponding to free energy change [Formula: see text] of 12.34 kcal mol( - 1), whereas the midpoint of the denaturation curve for T399A was approximately 3.94 M, corresponding to a [Formula: see text] of 4.45 kcal mol( - 1). Taken together, it can be concluded that the structural stability of BlGGT is superior to that of T399A.
Keywords: Autocatalytic processing; Bacillus licheniformis; Chemical denaturation; Thermal unfolding; γ-Glutamyltranspeptidase.
Figures



Similar articles
-
Experimental evidence for the involvement of amino acid residue Glu398 in the autocatalytic processing of Bacillus licheniformis γ-glutamyltranspeptidase.FEBS Open Bio. 2012 Sep 28;2:298-304. doi: 10.1016/j.fob.2012.09.007. Print 2012. FEBS Open Bio. 2012. PMID: 23772362 Free PMC article.
-
Biophysical characterization of Bacillus licheniformis and Escherichia coli γ-glutamyltranspeptidases: A comparative analysis.Int J Biol Macromol. 2011 Apr 1;48(3):414-22. doi: 10.1016/j.ijbiomac.2011.01.006. Epub 2011 Jan 14. Int J Biol Macromol. 2011. PMID: 21238482
-
Mutational Analysis of a Highly Conserved PLSSMXP Sequence in the Small Subunit of Bacillus licheniformis γ-Glutamyltranspeptidase.Biomolecules. 2019 Sep 19;9(9):508. doi: 10.3390/biom9090508. Biomolecules. 2019. PMID: 31546955 Free PMC article.
-
Characterization of glycine substitution mutations within the putative NAD+-binding site of Bacillus licheniformis aldehyde dehydrogenase.Protein Pept Lett. 2012 Nov;19(11):1183-93. doi: 10.2174/092986612803217097. Protein Pept Lett. 2012. PMID: 22587786
-
Enzymatic characterization of Bacillus licheniformis γ-glutamyltranspeptidase fused with N-terminally truncated forms of Bacillus sp. TS-23 α-amylase.Enzyme Microb Technol. 2012 Jul 15;51(2):86-94. doi: 10.1016/j.enzmictec.2012.04.005. Epub 2012 Apr 27. Enzyme Microb Technol. 2012. PMID: 22664192
Cited by
-
Bacterial Gamma-Glutamyl Transpeptidase, an Emerging Biocatalyst: Insights Into Structure-Function Relationship and Its Biotechnological Applications.Front Microbiol. 2021 Apr 9;12:641251. doi: 10.3389/fmicb.2021.641251. eCollection 2021. Front Microbiol. 2021. PMID: 33897647 Free PMC article. Review.
-
Experimental evidence for the involvement of amino acid residue Glu398 in the autocatalytic processing of Bacillus licheniformis γ-glutamyltranspeptidase.FEBS Open Bio. 2012 Sep 28;2:298-304. doi: 10.1016/j.fob.2012.09.007. Print 2012. FEBS Open Bio. 2012. PMID: 23772362 Free PMC article.
References
LinkOut - more resources
Full Text Sources

