Function and 3D structure of the N-glycans on glycoproteins
- PMID: 22942711
- PMCID: PMC3430242
- DOI: 10.3390/ijms13078398
Function and 3D structure of the N-glycans on glycoproteins
Abstract
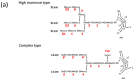
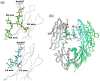
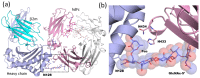
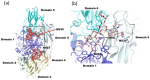
Glycosylation is one of the most common post-translational modifications in eukaryotic cells and plays important roles in many biological processes, such as the immune response and protein quality control systems. It has been notoriously difficult to study glycoproteins by X-ray crystallography since the glycan moieties usually have a heterogeneous chemical structure and conformation, and are often mobile. Nonetheless, recent technical advances in glycoprotein crystallography have accelerated the accumulation of 3D structural information. Statistical analysis of "snapshots" of glycoproteins can provide clues to understanding their structural and dynamic aspects. In this review, we provide an overview of crystallographic analyses of glycoproteins, in which electron density of the glycan moiety is clearly observed. These well-defined N-glycan structures are in most cases attributed to carbohydrate-protein and/or carbohydrate-carbohydrate interactions and may function as "molecular glue" to help stabilize inter- and intra-molecular interactions. However, the more mobile N-glycans on cell surface receptors, the electron density of which is usually missing on X-ray crystallography, seem to guide the partner ligand to its binding site and prevent irregular protein aggregation by covering oligomerization sites away from the ligand-binding site.
Keywords: N-glycan; glycoform; glycoprotein; protein crystallography.
Figures



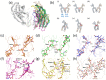


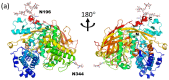
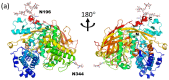

References
-
- Helenius A., Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. - PubMed
-
- Apweiler R., Hermjakob H., Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta. 1999;1473:4–8. - PubMed
-
- Nishikawa A., Ihara Y., Hatakeyama M., Kangawa K., Taniguchi N. Purification, cDNA cloning, and expression of UDP-N-acetylglucosamine:β-d-mannoside β-1,4-Nacetylglucosaminyltransferase III from rat kidney. J. Biol. Chem. 1992;267:18199–18204. - PubMed
-
- Cummings R.D., Trowbridge I.S., Kornfeld S. A mouse lymphoma cell line resistant to the leukoagglutinating lectin from Phaseolus vulgaris is deficient in UDP-GlcNAc:α-d-mannoside- β-1,6-N-acetylglucosaminyltransferase. J. Biol. Chem. 1982;257:13421–13427. - PubMed
-
- Uozumi N., Yanagidani S., Miyoshi E., Ihara Y., Sakuma T., Gao C.X., Teshima T., Fujii S., Shiba T., Taniguchi N. Purification and cDNA cloning of porcine brain GDP-l-Fuc:Nacetyl- β-d-glucosaminide α1→6 fucosyltransferase. J. Biol. Chem. 1996;271:27810–27817. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

