Protease-dependent mechanisms of complement evasion by bacterial pathogens
- PMID: 22944688
- PMCID: PMC3488274
- DOI: 10.1515/hsz-2012-0174
Protease-dependent mechanisms of complement evasion by bacterial pathogens
Abstract
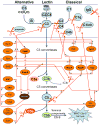
The human immune system has evolved a variety of mechanisms for the primary task of neutralizing and eliminating microbial intruders. As the first line of defense, the complement system is responsible for rapid recognition and opsonization of bacteria, presentation to phagocytes and bacterial cell killing by direct lysis. All successful human pathogens have mechanisms of circumventing the antibacterial activity of the complement system and escaping this stage of the immune response. One of the ways in which pathogens achieve this is the deployment of proteases. Based on the increasing number of recent publications in this area, it appears that proteolytic inactivation of the antibacterial activities of the complement system is a common strategy of avoiding targeting by this arm of host innate immune defense. In this review, we focus on those bacteria that deploy proteases capable of degrading complement system components into non-functional fragments, thus impairing complement-dependent antibacterial activity and facilitating pathogen survival inside the host.
Figures

Similar articles
-
Complement evasion strategies of pathogens-acquisition of inhibitors and beyond.Mol Immunol. 2009 Sep;46(14):2808-17. doi: 10.1016/j.molimm.2009.04.025. Epub 2009 May 27. Mol Immunol. 2009. PMID: 19477524 Review.
-
The hijackers guide to escaping complement: Lessons learned from pathogens.Mol Immunol. 2019 Oct;114:49-61. doi: 10.1016/j.molimm.2019.07.018. Epub 2019 Jul 20. Mol Immunol. 2019. PMID: 31336249 Review.
-
Utilizing complement evasion strategies to design complement-based antibacterial immunotherapeutics: Lessons from the pathogenic Neisseriae.Immunobiology. 2016 Oct;221(10):1110-23. doi: 10.1016/j.imbio.2016.05.016. Epub 2016 Jun 1. Immunobiology. 2016. PMID: 27297292 Free PMC article. Review.
-
Hijacking Factor H for Complement Immune Evasion.Front Immunol. 2021 Feb 25;12:602277. doi: 10.3389/fimmu.2021.602277. eCollection 2021. Front Immunol. 2021. PMID: 33717083 Free PMC article. Review.
-
The complement system.Cell Tissue Res. 2011 Jan;343(1):227-35. doi: 10.1007/s00441-010-1034-0. Epub 2010 Sep 14. Cell Tissue Res. 2011. PMID: 20838815 Free PMC article. Review.
Cited by
-
Evidence of mutualism between two periodontal pathogens: co-operative haem acquisition by the HmuY haemophore of Porphyromonas gingivalis and the cysteine protease interpain A (InpA) of Prevotella intermedia.Mol Oral Microbiol. 2013 Jun;28(3):219-29. doi: 10.1111/omi.12018. Epub 2013 Jan 22. Mol Oral Microbiol. 2013. PMID: 23336115 Free PMC article.
-
Salivary Total Protease Activity Based on a Broad-Spectrum Fluorescence Resonance Energy Transfer Approach to Monitor Induction and Resolution of Gingival Inflammation.Mol Diagn Ther. 2019 Oct;23(5):667-676. doi: 10.1007/s40291-019-00421-1. Mol Diagn Ther. 2019. PMID: 31372941 Free PMC article.
-
An Irreversible Inhibitor to Probe the Role of Streptococcus pyogenes Cysteine Protease SpeB in Evasion of Host Complement Defenses.ACS Chem Biol. 2020 Aug 21;15(8):2060-2069. doi: 10.1021/acschembio.0c00191. Epub 2020 Jul 28. ACS Chem Biol. 2020. PMID: 32662975 Free PMC article.
-
Porphyromonas gingivalis facilitates the development and progression of destructive arthritis through its unique bacterial peptidylarginine deiminase (PAD).PLoS Pathog. 2013 Sep;9(9):e1003627. doi: 10.1371/journal.ppat.1003627. Epub 2013 Sep 12. PLoS Pathog. 2013. PMID: 24068934 Free PMC article.
-
Peptidyl arginine deiminase from Porphyromonas gingivalis abolishes anaphylatoxin C5a activity.J Biol Chem. 2014 Nov 21;289(47):32481-7. doi: 10.1074/jbc.C114.617142. Epub 2014 Oct 16. J Biol Chem. 2014. PMID: 25324545 Free PMC article.
References
-
- Areschoug T, Stalhammar-Carlemalm M, Karlsson I, Lindahl G. Streptoccocal β protein has separate binding sites for human factor H and IgA-Fc. J Biol Chem. 2002;277:12642–12648. - PubMed
-
- Atkins KL, Burman JD, Chamberlain ES, Cooper JE, Poutrel B, Bagby S, Jenkins ATA, Feil EJ, Van den Elsen JMH. S. aureus IgG-binding proteins SpA and Sbi: host specificity and mechanisms of immune complex formation. Mol Immunol. 2008;45:1600–1611. - PubMed
-
- Banbula A, Potempa J, Travis J, Fernandez-Catala’n C, Mann K, Huber R, Bode W, Medrano F. Amino-acid sequence and three-dimensional structure of the Staphylococcus aureus metalloproteinase at 1.72Å resolution. Structure. 1998;6:1185–1193. - PubMed
-
- Bardoel BW, Strijp JA. Molecular battle between host and bacterium: recognition in innate immunity. J Mol Recognit. 2011;24:1077–86. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
