Coxsackievirus mutants that can bypass host factor PI4KIIIβ and the need for high levels of PI4P lipids for replication
- PMID: 22945356
- PMCID: PMC3494396
- DOI: 10.1038/cr.2012.129
Coxsackievirus mutants that can bypass host factor PI4KIIIβ and the need for high levels of PI4P lipids for replication
Abstract
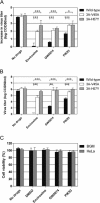
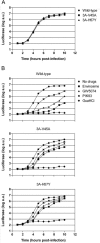
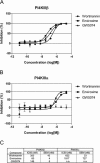
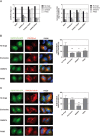
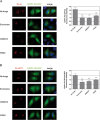
RNA viruses can rapidly mutate and acquire resistance to drugs that directly target viral enzymes, which poses serious problems in a clinical context. Therefore, there is a growing interest in the development of antiviral drugs that target host factors critical for viral replication, since they are unlikely to mutate in response to therapy. We recently demonstrated that phosphatidylinositol-4-kinase IIIβ (PI4KIIIβ) and its product phosphatidylinositol-4-phosphate (PI4P) are essential for replication of enteroviruses, a group of medically important RNA viruses including poliovirus (PV), coxsackievirus, rhinovirus, and enterovirus 71. Here, we show that enviroxime and GW5074 decreased PI4P levels at the Golgi complex by directly inhibiting PI4KIIIβ. Coxsackievirus mutants resistant to these inhibitors harbor single point mutations in the non-structural protein 3A. These 3A mutations did not confer compound-resistance by restoring the activity of PI4KIIIβ in the presence of the compounds. Instead, replication of the mutant viruses no longer depended on PI4KIIIβ, since their replication was insensitive to siRNA-mediated depletion of PI4KIIIβ. The mutant viruses also did not rely on other isoforms of PI4K. Consistently, no high level of PI4P could be detected at the replication sites induced by the mutant viruses in the presence of the compounds. Collectively, these findings indicate that through specific single point mutations in 3A, CVB3 can bypass an essential host factor and lipid for its propagation, which is a new example of RNA viruses acquiring resistance against antiviral compounds, even when they directly target host factors.
Figures
References
-
- Mallia P, Contoli M, Caramori G, Pandit A, Johnston SL, Papi A. Exacerbations of asthma and chronic obstructive pulmonary disease (COPD): focus on virus induced exacerbations. Curr Pharm Des. 2007;13:73–97. - PubMed
-
- Tebruegge M, Curtis N. Enterovirus infections in neonates. Semin Fetal Neonatal Med. 2009;14:222–227. - PubMed
-
- Whitton JL, Cornell CT, Feuer R. Host and virus determinants of picornavirus pathogenesis and tropism. Nat Rev Microbiol. 2005;3:765–776. - PubMed
-
- Bible JM, Pantelidis P, Chan PK, Tong CY. Genetic evolution of enterovirus 71: epidemiological and pathological implications. Rev Med Virol. 2007;17:371–379. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

