Age- and calorie restriction-related changes in rat brain mitochondrial DNA and TFAM binding
- PMID: 22945739
- PMCID: PMC3776104
- DOI: 10.1007/s11357-012-9465-z
Age- and calorie restriction-related changes in rat brain mitochondrial DNA and TFAM binding
Abstract
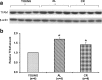
Aging markedly affects mitochondrial biogenesis and functions particularly in tissues highly dependent on the organelle's bioenergetics capability such as the brain's frontal cortex. Calorie restriction (CR) diet is, so far, the only intervention able to delay or prevent the onset of several age-related alterations in different organisms. We determined the contents of mitochondrial transcription factor A (TFAM), mitochondrial DNA (mtDNA), and the 4.8-kb mtDNA deletion in the frontal cortex from young (6-month-old) and aged (26-month-old), ad libitum-fed (AL) and calorie-restricted (CR), rats. We found a 70 % increase in TFAM amount, a 25 % loss in mtDNA content, and a 35 % increase in the 4.8-kb deletion content in the aged AL animals with respect to the young rats. TFAM-specific binding to six mtDNA regions was analyzed by mtDNA immunoprecipitation and semiquantitative polymerase chain reaction (PCR), showing a marked age-related decrease. Quantitative real-time PCR at two subregions involved in mtDNA replication demonstrated, in aged AL rats, a remarkable decrease (60-70 %) of TFAM-bound mtDNA. The decreased TFAM binding is a novel finding that may explain the mtDNA loss in spite of the compensatory TFAM increased amount. In aged CR rats, TFAM amount increased and mtDNA content decreased with respect to young rats' values, but the extent of the changes was smaller than in aged AL rats. Attenuation of the age-related effects due to the diet in the CR animals was further evidenced by the unchanged content of the 4.8-kb deletion with respect to that of young animals and by the partial prevention of the age-related decrease in TFAM binding to mtDNA.
Figures


References
-
- Aspnes LE, Lee CM, Weindruch R, Chung SS, Roecker EB, Aiken JM. Caloric restriction reduces fiber loss and mitochondrial abnormalities in aged rat muscle. FASEB J. 1997;11:573–581. - PubMed
-
- Bakala H, Delaval E, Hamelin M, Bismuth J, Borot-Laloi C, Corman B, Friguet B. Changes in rat liver mitochondria with aging. Lon protease-like reactivity and N(epsilon)-carboxymethyllysine accumulation in the matrix. Eur J Biochem. 2003;270:2295–2302. doi: 10.1046/j.1432-1033.2003.03598.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
