CYK4 inhibits Rac1-dependent PAK1 and ARHGEF7 effector pathways during cytokinesis
- PMID: 22945935
- PMCID: PMC3432774
- DOI: 10.1083/jcb.201204107
CYK4 inhibits Rac1-dependent PAK1 and ARHGEF7 effector pathways during cytokinesis
Abstract
In mitosis, animal cells lose their adhesion to the surrounding surfaces and become rounded. During mitotic exit, they reestablish these adhesions and at the same time physically contract and divide. How these competing processes are spatially segregated at the cell cortex remains mysterious. To address this question, we define the specific effector pathways used by RhoA and Rac1 in mitotic cells. We demonstrate that the MKlp1-CYK4 centralspindlin complex is a guanosine triphosphatase-activating protein (GAP) for Rac1 and not RhoA and that CYK4 negatively regulated Rac1 activity at the cell equator in anaphase. Cells expressing a CYK4 GAP mutant had defects in cytokinesis and showed elevated staining for the cell adhesion marker vinculin. These defects could be rescued by depletion of ARHGEF7 and p21-activated kinase, Rac1-specific effector proteins required for cell adhesion. Based on these findings, we propose that CYK4 GAP activity is required during anaphase to inhibit Rac1-dependent effector pathways associated with control of cell spreading and adhesion.
Figures
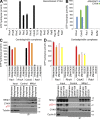
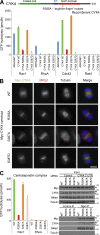
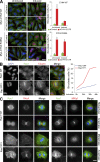

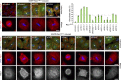
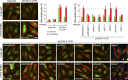
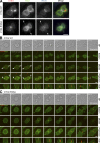
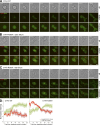
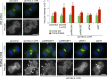
Comment in
-
Stuck in the middle: Rac, adhesion, and cytokinesis.J Cell Biol. 2012 Sep 3;198(5):769-71. doi: 10.1083/jcb.201207197. J Cell Biol. 2012. PMID: 22945931 Free PMC article.
References
-
- Castrillon D.H., Wasserman S.A. 1994. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 120:3367–3377 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

