Down-regulation of the IbEXP1 gene enhanced storage root development in sweetpotato
- PMID: 22945944
- PMCID: PMC3528024
- DOI: 10.1093/jxb/ers236
Down-regulation of the IbEXP1 gene enhanced storage root development in sweetpotato
Abstract
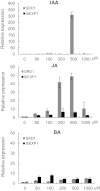
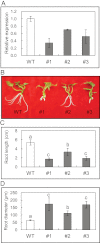
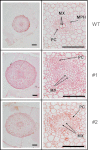
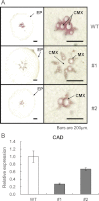
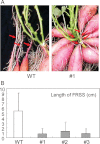
The role of an expansin gene (IbEXP1) in the formation of the storage root (SR) was investigated by expression pattern analysis and characterization of IbEXP1-antisense sweetpotato (Ipomoea batatas cv. Yulmi) plants in an attempt to elucidate the molecular mechanism underlying SR development in sweetpotato. The transcript level of IbEXP1 was high in the fibrous root (FR) and petiole at the FR stage, but decreased significantly at the young storage root (YSR) stage. IbEXP1-antisense plants cultured in vitro produced FRs which were both thicker and shorter than those of wild-type (WT) plants. Elongation growth of the epidermal cells was significantly reduced, and metaxylem and cambium cell proliferation was markedly enhanced in the FRs of IbEXP1-antisense plants, resulting in an earlier thickening growth in these plants relative to WT plants. There was a marked reduction in the lignification of the central stele of the FRs of the IbEXP1-antisense plants, suggesting that the FRs of the mutant plants possessed a higher potential than those of WT plants to develop into SRs. IbEXP1-antisense plants cultured in soil produced a larger number of SRs and, consequently, total SR weight per IbEXP1-antisense plant was greater than that per WT plant. These results demonstrate that SR development was accelerated in IbEXP1-antisense plants and suggest that IbEXP1 plays a negative role in the formation of SR by suppressing the proliferation of metaxylem and cambium cells to inhibit the initial thickening growth of SRs. IbEXP1 is the first sweetpotato gene whose role in SR development has been directly identified in soil-grown transgenic sweetpotato plants.
Figures



References
-
- Akita S, Yamamoto F, Ono M, Kusuhara M, Kobayashi H, Ikemoto S. 1962. Studies on the small tuber set method in sweetpotato cultivation. Bulletin of the Chugoku National Agricultural Experiment Station. 8, 75–128 (in Japanese with English summary)
-
- Belehu T, Hammes PS, Robbertse PJ. 2004. The origin and structure of adventitious roots in sweetpotato (Ipomoea batatas). Australian Journal of Botany. 52, 551–558
-
- Buchanan CD, Lim S, Salzman RA, et al. 2005. Sorghum bicolor’s transcriptome response to dehydration, high salinity and ABA. Plant Molecular Biology. 58, 699–720 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

