Using BAC transgenesis in zebrafish to identify regulatory sequences of the amyloid precursor protein gene in humans
- PMID: 22947103
- PMCID: PMC3546842
- DOI: 10.1186/1471-2164-13-451
Using BAC transgenesis in zebrafish to identify regulatory sequences of the amyloid precursor protein gene in humans
Abstract
Background: Non-coding DNA in and around the human Amyloid Precursor Protein (APP) gene that is central to Alzheimer's disease (AD) shares little sequence similarity with that of appb in zebrafish. Identifying DNA domains regulating expression of the gene in such situations becomes a challenge. Taking advantage of the zebrafish system that allows rapid functional analyses of gene regulatory sequences, we previously showed that two discontinuous DNA domains in zebrafish appb are important for expression of the gene in neurons: an enhancer in intron 1 and sequences 28-31 kb upstream of the gene. Here we identify the putative transcription factor binding sites responsible for this distal cis-acting regulation, and use that information to identify a regulatory region of the human APP gene.
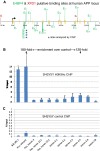
Results: Functional analyses of intron 1 enhancer mutations in enhancer-trap BACs expressed as transgenes in zebrafish identified putative binding sites of two known transcription factor proteins, E4BP4/ NFIL3 and Forkhead, to be required for expression of appb. A cluster of three E4BP4 sites at -31 kb is also shown to be essential for neuron-specific expression, suggesting that the dependence of expression on upstream sequences is mediated by these E4BP4 sites. E4BP4/ NFIL3 and XFD1 sites in the intron enhancer and E4BP4/ NFIL3 sites at -31 kb specifically and efficiently bind the corresponding zebrafish proteins in vitro. These sites are statistically over-represented in both the zebrafish appb and the human APP genes, although their locations are different. Remarkably, a cluster of four E4BP4 sites in intron 4 of human APP exists in actively transcribing chromatin in a human neuroblastoma cell-line, SHSY5Y, expressing APP as shown using chromatin immunoprecipitation (ChIP) experiments. Thus although the two genes share little sequence conservation, they appear to share the same regulatory logic and are regulated by a similar set of transcription factors.
Conclusion: The results suggest that the clock-regulated and immune system modulator transcription factor E4BP4/ NFIL3 likely regulates the expression of both appb in zebrafish and APP in humans. It suggests potential human APP gene regulatory pathways, not on the basis of comparing DNA primary sequences with zebrafish appb but on the model of conservation of transcription factors.
Figures




Similar articles
-
Harnessing mobile genetic elements to explore gene regulation.Mob Genet Elements. 2014 Jul 7;4:e29759. doi: 10.4161/mge.29759. eCollection 2014. Mob Genet Elements. 2014. PMID: 25054085 Free PMC article. Review.
-
Context dependent function of APPb enhancer identified using enhancer trap-containing BACs as transgenes in zebrafish.Nucleic Acids Res. 2008 Nov;36(19):6237-48. doi: 10.1093/nar/gkn628. Epub 2008 Oct 1. Nucleic Acids Res. 2008. PMID: 18832376 Free PMC article.
-
Generation of Alzheimer's Disease Transgenic Zebrafish Expressing Human APP Mutation Under Control of Zebrafish appb Promotor.Curr Alzheimer Res. 2017;14(6):668-679. doi: 10.2174/1567205013666161201202000. Curr Alzheimer Res. 2017. PMID: 27978793
-
Generation of transgenic zebrafish expressing green fluorescent protein under control of zebrafish amyloid precursor protein gene regulatory elements.Zebrafish. 2007 Winter;4(4):277-86. doi: 10.1089/zeb.2007.0516. Zebrafish. 2007. PMID: 18284334
-
A minireview of E4BP4/NFIL3 in heart failure.J Cell Physiol. 2018 Nov;233(11):8458-8466. doi: 10.1002/jcp.26790. Epub 2018 Jun 1. J Cell Physiol. 2018. PMID: 29856483 Review.
Cited by
-
Efficient site-specific transgenesis and enhancer activity tests in medaka using PhiC31 integrase.Development. 2013 Oct;140(20):4287-95. doi: 10.1242/dev.096081. Epub 2013 Sep 18. Development. 2013. PMID: 24048591 Free PMC article.
-
Effects of circadian clock genes and health-related behavior on metabolic syndrome in a Taiwanese population: Evidence from association and interaction analysis.PLoS One. 2017 Mar 15;12(3):e0173861. doi: 10.1371/journal.pone.0173861. eCollection 2017. PLoS One. 2017. PMID: 28296937 Free PMC article.
-
Identifying Distal cis-acting Gene-Regulatory Sequences by Expressing BACs Functionalized with loxP-Tn10 Transposons in Zebrafish.RSC Adv. 2013 Jun 21;3(23):8604-8617. doi: 10.1039/C3RA40332G. RSC Adv. 2013. PMID: 24772295 Free PMC article.
-
Harnessing mobile genetic elements to explore gene regulation.Mob Genet Elements. 2014 Jul 7;4:e29759. doi: 10.4161/mge.29759. eCollection 2014. Mob Genet Elements. 2014. PMID: 25054085 Free PMC article. Review.
-
A Human Embryonic Stem Cell Model of Aβ-Dependent Chronic Progressive Neurodegeneration.Front Neurosci. 2019 Sep 20;13:1007. doi: 10.3389/fnins.2019.01007. eCollection 2019. Front Neurosci. 2019. PMID: 31616241 Free PMC article.
References
-
- Dillen K, Annaert W. A two decade contribution of molecular cell biology to the centennial of Alzheimer’s disease: are we progressing toward therapy? Int Rev Cytol. 2006;254:215–300. - PubMed
-
- Theuns J, Brouwers N, Engelborghs S, Sleegers K, Bogaerts V, Corsmit E, De Pooter T, van Duijn CM, De Deyn PP, Van Broeckhoven C. Promoter mutations that increase amyloid precursor-protein expression are associated with Alzheimer disease. Am J Hum Genet. 2006;78:936–946. doi: 10.1086/504044. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

