Microarray analysis revealing common and distinct functions of promyelocytic leukemia protein (PML) and tumor necrosis factor alpha (TNFα) signaling in endothelial cells
- PMID: 22947142
- PMCID: PMC3542097
- DOI: 10.1186/1471-2164-13-453
Microarray analysis revealing common and distinct functions of promyelocytic leukemia protein (PML) and tumor necrosis factor alpha (TNFα) signaling in endothelial cells
Abstract
Background: Promyelocytic leukemia protein (PML) is a tumor suppressor that is highly expressed in endothelial cells nonetheless its role in endothelial cell biology remains elusive. Tumor necrosis factor alpha (TNFα) is an important cytokine associated with many inflammation-related diseases. We have previously demonstrated that TNFα induces PML protein accumulation. We hypothesized that PML may play a role in TNFα signaling pathway. To identify potential PML target genes and investigate the putative crosstalk between PML's function and TNFα signaling in endothelial cells, we carried out a microarray analysis in human primary umbilical endothelial cells (HUVECs).
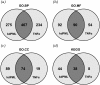
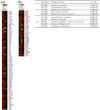
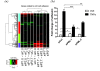
Results: We found that PML and TNFα regulate common and distinct genes involved in a similar spectrum of biological processes, pathways and human diseases. More importantly, we found that PML is required for fine-tuning of TNFα-mediated immune and inflammatory responses. Furthermore, our data suggest that PML and TNFα synergistically regulate cell adhesion by engaging multiple molecular mechanisms. Our biological functional assays exemplified that adhesion of U937 human leukocytes to HUVECs is co-regulated by PML and TNFα signaling.
Conclusions: Together, our study identified PML as an essential regulator of TNFα signaling by revealing the crosstalk between PML knockdown-mediated effects and TNFα-elicited signaling, thereby providing novel insights into TNFα signaling in endothelial cells.
Figures


Similar articles
-
Promyelocytic leukemia protein (PML) regulates endothelial cell network formation and migration in response to tumor necrosis factor α (TNFα) and interferon α (IFNα).J Biol Chem. 2012 Jul 6;287(28):23356-67. doi: 10.1074/jbc.M112.340505. Epub 2012 May 15. J Biol Chem. 2012. PMID: 22589541 Free PMC article.
-
Translational control of PML contributes to TNFα-induced apoptosis of MCF7 breast cancer cells and decreased angiogenesis in HUVECs.Cell Death Differ. 2016 Mar;23(3):469-83. doi: 10.1038/cdd.2015.114. Epub 2015 Sep 18. Cell Death Differ. 2016. PMID: 26383972 Free PMC article.
-
Histone deacetylase 7 promotes PML sumoylation and is essential for PML nuclear body formation.Mol Cell Biol. 2008 Sep;28(18):5658-67. doi: 10.1128/MCB.00874-08. Epub 2008 Jul 14. Mol Cell Biol. 2008. PMID: 18625722 Free PMC article.
-
Targeting promyelocytic leukemia protein: a means to regulating PML nuclear bodies.Int J Biol Sci. 2009 May 22;5(4):366-76. doi: 10.7150/ijbs.5.366. Int J Biol Sci. 2009. PMID: 19471587 Free PMC article. Review.
-
Cytoplasmic PML: from molecular regulation to biological functions.J Cell Biochem. 2014 May;115(5):812-8. doi: 10.1002/jcb.24727. J Cell Biochem. 2014. PMID: 24288198 Review.
Cited by
-
Interleukin-1β Stimulates Matrix Metalloproteinase 10 Secretion: A Possible Mechanism in Trophoblast-Dependent Spiral Artery Remodeling.FASEB J. 2025 May 15;39(9):e70597. doi: 10.1096/fj.202402329RR. FASEB J. 2025. PMID: 40326797 Free PMC article.
-
BLAT2DOLite: An Online System for Identifying Significant Relationships between Genetic Sequences and Diseases.PLoS One. 2016 Jun 17;11(6):e0157274. doi: 10.1371/journal.pone.0157274. eCollection 2016. PLoS One. 2016. PMID: 27315278 Free PMC article.
-
Promyelocytic leukemia protein regulates angiogenesis and epithelial-mesenchymal transition to limit metastasis in MDA-MB-231 breast cancer cells.Mol Oncol. 2023 Oct;17(10):2090-2108. doi: 10.1002/1878-0261.13501. Epub 2023 Sep 4. Mol Oncol. 2023. PMID: 37518985 Free PMC article.
-
The PML1-WDR5 axis regulates H3K4me3 marks and promotes stemness of estrogen receptor-positive breast cancer.Cell Death Differ. 2024 Jun;31(6):768-778. doi: 10.1038/s41418-024-01294-6. Epub 2024 Apr 16. Cell Death Differ. 2024. PMID: 38627584 Free PMC article.
-
β-Transducin repeat-containing protein 1 (β-TrCP1)-mediated silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) protein degradation promotes tumor necrosis factor α (TNFα)-induced inflammatory gene expression.J Biol Chem. 2013 Aug 30;288(35):25375-25386. doi: 10.1074/jbc.M113.473124. Epub 2013 Jul 16. J Biol Chem. 2013. PMID: 23861398 Free PMC article.
References
-
- Terris B, Baldin V, Dubois S, Degott C, Flejou JF, Henin D, Dejean A. PML nuclear bodies are general targets for inflammation and cell proliferation. Cancer res. 1995;55:1590–1597. - PubMed
-
- Crowder C, Dahle O, Davis RE, Gabrielsen OS, Rudikoff S. PML mediates IFN-alpha-induced apoptosis in myeloma by regulating TRAIL induction. Blood. 2005;105:1280–1287. - PubMed
-
- Kakizuka A, Miller WHJ, Umesono K, Warrell RPJ, Frankel SR, Murty VV, Dmitrovsky E, Evans RM. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-C. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

