Calmodulin binds a highly extended HIV-1 MA protein that refolds upon its release
- PMID: 22947870
- PMCID: PMC3414894
- DOI: 10.1016/j.bpj.2012.06.042
Calmodulin binds a highly extended HIV-1 MA protein that refolds upon its release
Erratum in
-
Calmodulin Binds a Highly Extended HIV-1 MA Protein That Refolds Upon Its Release.Biophys J. 2023 May 2;122(9):1734. doi: 10.1016/j.bpj.2023.04.004. Epub 2023 Apr 8. Biophys J. 2023. PMID: 37031681 Free PMC article. No abstract available.
Abstract
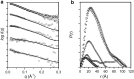
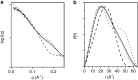
Calmodulin (CaM) expression is upregulated upon HIV-1 infection and interacts with proteins involved in viral processing, including the multifunctional HIV-1 MA protein. We present here the results of studies utilizing small-angle neutron scattering with contrast variation that, when considered in the light of earlier fluorescence and NMR data, show CaM binds MA in an extended open-clamp conformation via interactions with two tryptophans that are widely spaced in sequence and space. The interaction requires a disruption of the MA tertiary fold such that MA becomes highly extended in a long snakelike conformation. The CaM-MA interface is extensive, covering ~70% of the length of the MA such that regions known to be important in MA interactions with critical binding partners would be impacted. The CaM conformation is semiextended and as such is distinct from the classical CaM-collapse about short α-helical targets. NMR data show that upon dissociation of the CaM-MA complex, either by the removal of Ca2+ or increasing ionic strength, MA reforms its native tertiary contacts. Thus, we observe a high level of structural plasticity in MA that may facilitate regulation of its activities via intracellular Ca2+-signaling during viral processing.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Solution structure of calmodulin bound to the binding domain of the HIV-1 matrix protein.J Biol Chem. 2014 Mar 21;289(12):8697-705. doi: 10.1074/jbc.M113.543694. Epub 2014 Feb 5. J Biol Chem. 2014. PMID: 24500712 Free PMC article.
-
Calmodulin disrupts the structure of the HIV-1 MA protein.J Mol Biol. 2010 Jul 23;400(4):702-14. doi: 10.1016/j.jmb.2010.05.022. Epub 2010 May 19. J Mol Biol. 2010. PMID: 20488189 Free PMC article.
-
NMR, biophysical, and biochemical studies reveal the minimal Calmodulin binding domain of the HIV-1 matrix protein.J Biol Chem. 2011 Sep 23;286(38):33533-43. doi: 10.1074/jbc.M111.273623. Epub 2011 Jul 28. J Biol Chem. 2011. PMID: 21799007 Free PMC article.
-
The Interplay between HIV-1 Gag Binding to the Plasma Membrane and Env Incorporation.Viruses. 2020 May 16;12(5):548. doi: 10.3390/v12050548. Viruses. 2020. PMID: 32429351 Free PMC article. Review.
-
Roles played by acidic lipids in HIV-1 Gag membrane binding.Virus Res. 2014 Nov 26;193:108-15. doi: 10.1016/j.virusres.2014.06.015. Epub 2014 Jul 3. Virus Res. 2014. PMID: 24998886 Free PMC article. Review.
Cited by
-
Insights into open/closed conformations of the catalytically active human guanylate kinase as investigated by small-angle X-ray scattering.Eur Biophys J. 2016 Jan;45(1):81-9. doi: 10.1007/s00249-015-1079-9. Epub 2015 Oct 7. Eur Biophys J. 2016. PMID: 26446352 Free PMC article.
-
Calmodulin extracts the Ras family protein RalA from lipid bilayers by engagement with two membrane-targeting motifs.Proc Natl Acad Sci U S A. 2021 Sep 7;118(36):e2104219118. doi: 10.1073/pnas.2104219118. Proc Natl Acad Sci U S A. 2021. PMID: 34480001 Free PMC article.
-
Solution structure of calmodulin bound to the binding domain of the HIV-1 matrix protein.J Biol Chem. 2014 Mar 21;289(12):8697-705. doi: 10.1074/jbc.M113.543694. Epub 2014 Feb 5. J Biol Chem. 2014. PMID: 24500712 Free PMC article.
-
Comparative Genomics of Amphibian-like Ranaviruses, Nucleocytoplasmic Large DNA Viruses of Poikilotherms.Evol Bioinform Online. 2016 Oct 25;11(Suppl 2):71-82. doi: 10.4137/EBO.S33490. eCollection 2015. Evol Bioinform Online. 2016. PMID: 27812275 Free PMC article.
-
Bayesian inference of protein conformational ensembles from limited structural data.PLoS Comput Biol. 2018 Dec 17;14(12):e1006641. doi: 10.1371/journal.pcbi.1006641. eCollection 2018 Dec. PLoS Comput Biol. 2018. PMID: 30557358 Free PMC article.
References
-
- Chin D., Means A.R. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. - PubMed
-
- Yamniuk A.P., Vogel H.J. Calmodulin's flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 2004;27:33–57. - PubMed
-
- Miller M.A., Mietzner T.A., et al. Montelaro R.C. Identification of a calmodulin-binding and inhibitory peptide domain in the HIV-1 transmembrane glycoprotein. AIDS Res. Hum. Retroviruses. 1993;9:1057–1066. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

