Rapid calculation of protein pKa values using Rosetta
- PMID: 22947875
- PMCID: PMC3414879
- DOI: 10.1016/j.bpj.2012.06.044
Rapid calculation of protein pKa values using Rosetta
Abstract
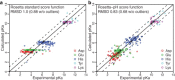
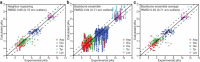
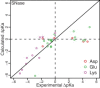
We developed a Rosetta-based Monte Carlo method to calculate the pK(a) values of protein residues that commonly exhibit variable protonation states (Asp, Glu, Lys, His, and Tyr). We tested the technique by calculating pK(a) values for 264 residues from 34 proteins. The standard Rosetta score function, which is independent of any environmental conditions, failed to capture pK(a) shifts. After incorporating a Coulomb electrostatic potential and optimizing the solvation reference energies for pK(a) calculations, we employed a method that allowed side-chain flexibility and achieved a root mean-square deviation (RMSD) of 0.83 from experimental values (0.68 after discounting 11 predictions with an error over 2 pH units). Additional degrees of side-chain conformational freedom for the proximal residues facilitated the capture of charge-charge interactions in a few cases, resulting in an overall RMSD of 0.85 pH units. The addition of backbone flexibility increased the overall RMSD to 0.93 pH units but improved relative pK(a) predictions for proximal catalytic residues. The method also captures large pK(a) shifts of lysine and some glutamate point mutations in staphylococcal nuclease. Thus, a simple and fast method based on the Rosetta score function and limited conformational sampling produces pK(a) values that will be useful when rapid estimation is essential, such as in docking, design, and folding.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
A fast and simple method to calculate protonation states in proteins.Proteins. 1999 Sep 1;36(4):474-83. doi: 10.1002/(sici)1097-0134(19990901)36:4<474::aid-prot12>3.0.co;2-v. Proteins. 1999. PMID: 10450090
-
Exploring conformational changes coupled to ionization states using a hybrid Rosetta-MCCE protocol.Proteins. 2011 Dec;79(12):3356-63. doi: 10.1002/prot.23146. Epub 2011 Aug 30. Proteins. 2011. PMID: 22072519
-
Calbindin D(9k): a protein optimized for calcium binding at neutral pH.Biochemistry. 2001 Dec 18;40(50):15334-40. doi: 10.1021/bi0114022. Biochemistry. 2001. PMID: 11735416
-
The pKa Cooperative: a collaborative effort to advance structure-based calculations of pKa values and electrostatic effects in proteins.Proteins. 2011 Dec;79(12):3249-59. doi: 10.1002/prot.23194. Epub 2011 Oct 15. Proteins. 2011. PMID: 22002877 Free PMC article. Review.
-
Progress in the prediction of pKa values in proteins.Proteins. 2011 Dec;79(12):3260-75. doi: 10.1002/prot.23189. Epub 2011 Oct 15. Proteins. 2011. PMID: 22002859 Free PMC article. Review.
Cited by
-
The first crystal structure of human RNase 6 reveals a novel substrate-binding and cleavage site arrangement.Biochem J. 2016 Jun 1;473(11):1523-36. doi: 10.1042/BCJ20160245. Epub 2016 Mar 24. Biochem J. 2016. PMID: 27013146 Free PMC article.
-
Cholesterol-induced suppression of Kir2 channels is mediated by decoupling at the inter-subunit interfaces.iScience. 2022 Apr 29;25(5):104329. doi: 10.1016/j.isci.2022.104329. eCollection 2022 May 20. iScience. 2022. PMID: 35602957 Free PMC article.
-
Structural Dynamics of the Lipid Antigen-Binding Site of CD1d Protein.Biomolecules. 2020 Apr 1;10(4):532. doi: 10.3390/biom10040532. Biomolecules. 2020. PMID: 32244759 Free PMC article.
-
IPC 2.0: prediction of isoelectric point and pKa dissociation constants.Nucleic Acids Res. 2021 Jul 2;49(W1):W285-W292. doi: 10.1093/nar/gkab295. Nucleic Acids Res. 2021. PMID: 33905510 Free PMC article.
-
Experimental pKa Value Determination of All Ionizable Groups of a Hyperstable Protein.Chembiochem. 2019 Apr 1;20(7):922-930. doi: 10.1002/cbic.201800628. Epub 2019 Feb 11. Chembiochem. 2019. PMID: 30511779 Free PMC article.
References
-
- Sheinerman F.B., Norel R., Honig B. Electrostatic aspects of protein-protein interactions. Curr. Opin. Struct. Biol. 2000;10:153–159. - PubMed
-
- Warshel A., Dryga A. Simulating electrostatic energies in proteins: perspectives and some recent studies of pKas, redox, and other crucial functional properties. Proteins. 2011;79:3469–3484. - PubMed
-
- Warren G.L., Andrews C.W., Head M.S. A critical assessment of docking programs and scoring functions. J. Med. Chem. 2006;49:5912–5931. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

