Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors
- PMID: 22948769
- PMCID: PMC3460180
- DOI: 10.1101/gr.137976.112
Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors
Abstract
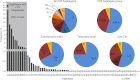
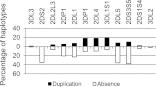
The KIR complex appears to be evolving rapidly in humans, and more than 50 different haplotypes have been described, ranging from four to 14 KIR loci. Previously it has been suggested that most KIR haplotypes consist of framework genes, present in all individuals, which bracket a variable number of other genes. We used a new technique to type 793 families from the United Kingdom and United States for both the presence/absence of all individual KIR genes as well as copy number and found that KIR haplotypes are even more complex. It is striking that all KIR loci are subject to copy number variation (CNV), including the so-called framework genes, but CNV is much more frequent in KIR B haplotypes than KIR A haplotypes. These two basic KIR haplotype groups, A and B, appear to be following different evolutionary trajectories. Despite the great diversity, there are 11 common haplotypes, derived by reciprocal recombination near KIR2DL4, which collectively account for 94% of KIR haplotypes determined in Caucasian samples. These haplotypes could be derived from combinations of just three centromeic and two telomeric motifs, simplifying disease analysis for these haplotypes. The remaining 6% of haplotypes displayed novel examples of expansion and contraction of numbers of loci. Conventional KIR typing misses much of this additional complexity, with important implications for studying the genetics of disease association with KIR that can now be explored by CNV analysis.
Figures




References
-
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR 2002. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30: 97–101 - PubMed
-
- Gomez-Lozano N, Gardiner CM, Parham P, Vilches C 2002. Some human KIR haplotypes contain two KIR2DL5 genes: KIR2DL5A and KIR2DL5B. Immunogenetics 54: 314–319 - PubMed
-
- Gomez-Lozano N, de Pablo R, Puente S, Vilches C 2003. Recognition of HLA-G by the NK cell receptor KIR2DL4 is not essential for human reproduction. Eur J Immunol 33: 639–644 - PubMed
-
- Gomez-Lozano N, Estefania E, Williams F, Halfpenny I, Middleton D, Solis R, Vilches C 2005. The silent KIR3DP1 gene (CD158c) is transcribed and might encode a secreted receptor in a minority of humans, in whom the KIR3DP1, KIR2DL4 and KIR3DL1/KIR3DS1 genes are duplicated. Eur J Immunol 35: 16–24 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
