Chemical methods to interrogate bacterial quorum sensing pathways
- PMID: 22948815
- PMCID: PMC3480174
- DOI: 10.1039/c2ob26353j
Chemical methods to interrogate bacterial quorum sensing pathways
Abstract
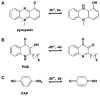
Bacteria frequently manifest distinct phenotypes as a function of cell density in a phenomenon known as quorum sensing (QS). This intercellular signalling process is mediated by "chemical languages" comprised of low-molecular weight signals, known as autoinducers, and their cognate receptor proteins. As many of the phenotypes regulated by QS can have a significant impact on the success of pathogenic or mutualistic prokaryotic-eukaryotic interactions, there is considerable interest in methods to probe and modulate QS pathways with temporal and spatial control. Such methods would be valuable for both basic research in bacterial ecology and in practical medicinal, agricultural, and industrial applications. Toward this goal, considerable recent research has been focused on the development of chemical approaches to study bacterial QS pathways. In this Perspective, we provide an overview of the use of chemical probes and techniques in QS research. Specifically, we focus on: (1) combinatorial approaches for the discovery of small molecule QS modulators, (2) affinity chromatography for the isolation of QS receptors, (3) reactive and fluorescent probes for QS receptors, (4) antibodies as quorum "quenchers," (5) abiotic polymeric "sinks" and "pools" for QS signals, and (6) the electrochemical sensing of QS signals. The application of such chemical methods can offer unique advantages for both elucidating and manipulating QS pathways in culture and under native conditions.
Figures


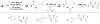

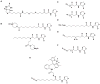
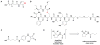

Similar articles
-
Non-native N-aroyl L-homoserine lactones are potent modulators of the quorum sensing receptor RpaR in Rhodopseudomonas palustris.Chembiochem. 2014 Jan 3;15(1):87-93. doi: 10.1002/cbic.201300570. Epub 2013 Nov 26. Chembiochem. 2014. PMID: 24281952 Free PMC article.
-
Mathematical Modelling of Bacterial Quorum Sensing: A Review.Bull Math Biol. 2016 Aug;78(8):1585-639. doi: 10.1007/s11538-016-0160-6. Epub 2016 Aug 25. Bull Math Biol. 2016. PMID: 27561265 Review.
-
Production, detection and application perspectives of quorum sensing autoinducer-2 in bacteria.J Biotechnol. 2018 Feb 20;268:53-60. doi: 10.1016/j.jbiotec.2018.01.009. Epub 2018 Jan 31. J Biotechnol. 2018. PMID: 29355813 Review.
-
Bacterial quorum sensing in symbiotic and pathogenic relationships with hosts.Biosci Biotechnol Biochem. 2018 Mar;82(3):363-371. doi: 10.1080/09168451.2018.1433992. Epub 2018 Feb 9. Biosci Biotechnol Biochem. 2018. PMID: 29424268 Review.
-
Recent progresses on AI-2 bacterial quorum sensing inhibitors.Curr Med Chem. 2012;19(2):174-86. doi: 10.2174/092986712803414187. Curr Med Chem. 2012. PMID: 22320296 Review.
Cited by
-
Natural Products as Platforms To Overcome Antibiotic Resistance.Chem Rev. 2017 Oct 11;117(19):12415-12474. doi: 10.1021/acs.chemrev.7b00283. Epub 2017 Sep 27. Chem Rev. 2017. PMID: 28953368 Free PMC article. Review.
-
A Comparative Analysis of Acyl-Homoserine Lactone Synthase Assays.Chembiochem. 2015 Dec;16(18):2651-9. doi: 10.1002/cbic.201500387. Epub 2015 Nov 6. Chembiochem. 2015. PMID: 26456773 Free PMC article.
-
Small molecule disruption of quorum sensing cross-regulation in pseudomonas aeruginosa causes major and unexpected alterations to virulence phenotypes.J Am Chem Soc. 2015 Feb 4;137(4):1510-9. doi: 10.1021/ja5110798. Epub 2015 Jan 22. J Am Chem Soc. 2015. PMID: 25574853 Free PMC article.
-
A Comparative Analysis of Synthetic Quorum Sensing Modulators in Pseudomonas aeruginosa: New Insights into Mechanism, Active Efflux Susceptibility, Phenotypic Response, and Next-Generation Ligand Design.J Am Chem Soc. 2015 Nov 25;137(46):14626-39. doi: 10.1021/jacs.5b06728. Epub 2015 Nov 11. J Am Chem Soc. 2015. PMID: 26491787 Free PMC article.
-
Phenazine Antibiotic-Inspired Discovery of Bacterial Biofilm-Eradicating Agents.Chembiochem. 2019 Dec 2;20(23):2885-2902. doi: 10.1002/cbic.201900116. Epub 2019 Oct 2. Chembiochem. 2019. PMID: 30811834 Free PMC article. Review.
References
-
- Galloway WRJD, Hodgkinson JT, Bowden SD, Welch M, Spring DR. Chem Rev. 2011;111:28–67. - PubMed
-
- Uroz S, Dessaux Y, Oger P. ChemBioChem. 2009;10:205–216. - PubMed
-
- Bauer WD, Mathesius U. Curr Opin Plant Biol. 2004;7:429–433. - PubMed
-
- Teplitski M, Mathesius U, Rumbaugh KP. Chem Rev. 2011;111:100–116. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

