The dynamics of DNA methylation in schizophrenia and related psychiatric disorders
- PMID: 22948975
- PMCID: PMC3521968
- DOI: 10.1038/npp.2012.125
The dynamics of DNA methylation in schizophrenia and related psychiatric disorders
Abstract
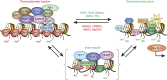
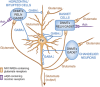
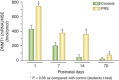
Major psychiatric disorders such as schizophrenia (SZ) and bipolar disorder (BP) with psychosis (BP+) express a complex symptomatology characterized by positive symptoms, negative symptoms, and cognitive impairment. Postmortem studies of human SZ and BP+ brains show considerable alterations in the transcriptome of a variety of cortical structures, including multiple mRNAs that are downregulated in both inhibitory GABAergic and excitatory pyramidal neurons compared with non-psychiatric subjects (NPS). Several reports show increased expression of DNA methyltransferases in telencephalic GABAergic neurons. Accumulating evidence suggests a critical role for altered DNA methylation processes in the pathogenesis of SZ and related psychiatric disorders. The establishment and maintenance of CpG site methylation is essential during central nervous system differentiation and this methylation has been implicated in synaptic plasticity, learning, and memory. Atypical hypermethylation of candidate gene promoters expressed in GABAergic neurons is associated with transcriptional downregulation of the corresponding mRNAs, including glutamic acid decarboxylase 67 (GAD67) and reelin (RELN). Recent reports indicate that the methylation status of promoter proximal CpG dinucleotides is in a dynamic balance between DNA methylation and DNA hydroxymethylation. Hydroxymethylation and subsequent DNA demethylation is more complex and involves additional proteins downstream of 5-hydroxymethylcytosine, including members of the base excision repair (BER) pathway. Recent advances in our understanding of altered CpG methylation, hydroxymethylation, and active DNA demethylation provide a framework for the identification of new targets, which may be exploited for the pharmacological intervention of the psychosis associated with SZ and possibly BP+.
Figures


References
-
- Aapola U, Kawasaki K, Scott HS, Ollila J, Vihinen M, Heino M, et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65:293–298. - PubMed
-
- Aapola U, Lyle R, Krohn K, Antonarakis SE, Peterson P. Isolationand initial characterization of the mouse Dnmt3l gene. Cytogenet Cell Genet. 2001;92:122–126. - PubMed
-
- Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, et al. 2005Hypermethylation of the reelin (RELN). promoter in the brain of schizophrenic patients: a preliminary report Am J Med Genet B Neuropsychiatr Genet 13460–66.The first paper to provide biochemical evidence that the reelin promoter is hypermethylated in the brain of schizophrenia patients. - PubMed
-
- Abdolmaleky HM, Thiagalingam S. Can the schizophrenia epigenome provide clues for the molecular basis of pathogenesis. Epigenomics. 2011;3:679–683. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

