Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering
- PMID: 22949640
- PMCID: PMC3465404
- DOI: 10.1073/pnas.1213813109
Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering
Abstract
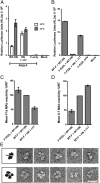
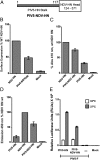
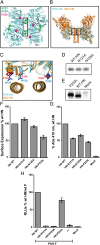
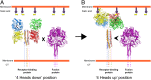
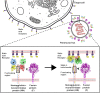
The Paramyxoviridae family of enveloped viruses enters cells through the concerted action of two viral glycoproteins. The receptor-binding protein, hemagglutinin-neuraminidase (HN), H, or G, binds its cellular receptor and activates the fusion protein, F, which, through an extensive refolding event, brings viral and cellular membranes together, mediating virus-cell fusion. However, the underlying mechanism of F activation on receptor engagement remains unclear. Current hypotheses propose conformational changes in HN, H, or G propagating from the receptor-binding site in the HN, H, or G globular head to the F-interacting stalk region. We provide evidence that the receptor-binding globular head domain of the paramyxovirus parainfluenza virus 5 HN protein is entirely dispensable for F activation. Considering together the crystal structures of HN from different paramyxoviruses, varying energy requirements for fusion activation, F activation involving the parainfluenza virus 5 HN stalk domain, and properties of a chimeric paramyxovirus HN protein, we propose a simple model for the activation of paramyxovirus fusion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Heminway BR, Yu Y, Galinski MS. Paramyxovirus mediated cell fusion requires co-expression of both the fusion and hemagglutinin-neuraminidase glycoproteins. Virus Res. 1994;31:1–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

