Epistasis dominates the genetic architecture of Drosophila quantitative traits
- PMID: 22949659
- PMCID: PMC3465439
- DOI: 10.1073/pnas.1213423109
Epistasis dominates the genetic architecture of Drosophila quantitative traits
Abstract
Epistasis-nonlinear genetic interactions between polymorphic loci-is the genetic basis of canalization and speciation, and epistatic interactions can be used to infer genetic networks affecting quantitative traits. However, the role that epistasis plays in the genetic architecture of quantitative traits is controversial. Here, we compared the genetic architecture of three Drosophila life history traits in the sequenced inbred lines of the Drosophila melanogaster Genetic Reference Panel (DGRP) and a large outbred, advanced intercross population derived from 40 DGRP lines (Flyland). We assessed allele frequency changes between pools of individuals at the extremes of the distribution for each trait in the Flyland population by deep DNA sequencing. The genetic architecture of all traits was highly polygenic in both analyses. Surprisingly, none of the SNPs associated with the traits in Flyland replicated in the DGRP and vice versa. However, the majority of these SNPs participated in at least one epistatic interaction in the DGRP. Despite apparent additive effects at largely distinct loci in the two populations, the epistatic interactions perturbed common, biologically plausible, and highly connected genetic networks. Our analysis underscores the importance of epistasis as a principal factor that determines variation for quantitative traits and provides a means to uncover genetic networks affecting these traits. Knowledge of epistatic networks will contribute to our understanding of the genetic basis of evolutionarily and clinically important traits and enhance predictive ability at an individualized level in medicine and agriculture.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
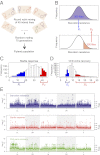

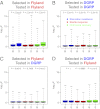

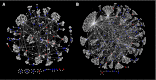
References
-
- Hindorff LA, et al. A Catalog of Published Genome-Wide Association Studies. Available at www.genome.gov/gwastudies. Accessed May 8, 2012.
-
- Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. 4th Ed. Essex, England: Addison Wesley Longman; 1996.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

