Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3'-terminal phosphate and 5'-OH
- PMID: 22949672
- PMCID: PMC3458315
- DOI: 10.1073/pnas.1213795109
Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3'-terminal phosphate and 5'-OH
Abstract
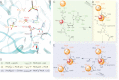
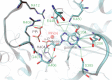
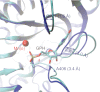
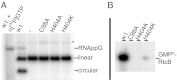
The RtcB protein has recently been identified as a 3'-phosphate RNA ligase that directly joins an RNA strand ending with a 2',3'-cyclic phosphate to the 5'-hydroxyl group of another RNA strand in a GTP/Mn(2+)-dependent reaction. Here, we report two crystal structures of Pyrococcus horikoshii RNA-splicing ligase RtcB in complex with Mn(2+) alone (RtcB/ Mn(2+)) and together with a covalently bound GMP (RtcB-GMP/Mn(2+)). The RtcB/ Mn(2+) structure (at 1.6 Å resolution) shows two Mn(2+) ions at the active site, and an array of sulfate ions nearby that indicate the binding sites of the RNA phosphate backbone. The structure of the RtcB-GMP/Mn(2+) complex (at 2.3 Å resolution) reveals the detailed geometry of guanylylation of histidine 404. The critical roles of the key residues involved in the binding of the two Mn(2+) ions, the four sulfates, and GMP are validated in extensive mutagenesis and biochemical experiments, which also provide a thorough characterization for the three steps of the RtcB ligation pathway: (i) guanylylation of the enzyme, (ii) guanylyl-transfer to the RNA substrate, and (iii) overall ligation. These results demonstrate that the enzyme's substrate-induced GTP binding site and the putative reactive RNA ends are in the vicinity of the binuclear Mn(2+) active center, which provides detailed insight into how the enzyme-bound GMP is tansferred to the 3'-phosphate of the RNA substrate for activation and subsequent nucleophilic attack by the 5'-hydroxyl of the second RNA substrate, resulting in the ligated product and release of GMP.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
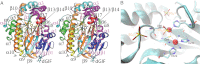
Similar articles
-
Structures of the noncanonical RNA ligase RtcB reveal the mechanism of histidine guanylylation.Biochemistry. 2013 Apr 16;52(15):2518-25. doi: 10.1021/bi4002375. Epub 2013 Apr 5. Biochemistry. 2013. PMID: 23560983 Free PMC article.
-
Distinct Contributions of Enzymic Functional Groups to the 2',3'-Cyclic Phosphodiesterase, 3'-Phosphate Guanylylation, and 3'-ppG/5'-OH Ligation Steps of the Escherichia coli RtcB Nucleic Acid Splicing Pathway.J Bacteriol. 2016 Mar 31;198(8):1294-304. doi: 10.1128/JB.00913-15. Print 2016 Apr. J Bacteriol. 2016. PMID: 26858100 Free PMC article.
-
The sequential 2',3'-cyclic phosphodiesterase and 3'-phosphate/5'-OH ligation steps of the RtcB RNA splicing pathway are GTP-dependent.Nucleic Acids Res. 2012 Sep 1;40(17):8558-67. doi: 10.1093/nar/gks558. Epub 2012 Jun 22. Nucleic Acids Res. 2012. PMID: 22730297 Free PMC article.
-
Insights into the structure and function of the RNA ligase RtcB.Cell Mol Life Sci. 2023 Nov 7;80(12):352. doi: 10.1007/s00018-023-05001-5. Cell Mol Life Sci. 2023. PMID: 37935993 Free PMC article. Review.
-
The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases.Curr Opin Struct Biol. 2004 Dec;14(6):757-64. doi: 10.1016/j.sbi.2004.10.006. Curr Opin Struct Biol. 2004. PMID: 15582400 Review.
Cited by
-
A tRNA splicing operon: Archease endows RtcB with dual GTP/ATP cofactor specificity and accelerates RNA ligation.Nucleic Acids Res. 2014 Apr;42(6):3931-42. doi: 10.1093/nar/gkt1375. Epub 2014 Jan 16. Nucleic Acids Res. 2014. PMID: 24435797 Free PMC article.
-
Fungi of the order Mucorales express a "sealing-only" tRNA ligase.RNA. 2024 Mar 18;30(4):354-366. doi: 10.1261/rna.079957.124. RNA. 2024. PMID: 38307611 Free PMC article.
-
Distinctive kinetics and substrate specificities of plant and fungal tRNA ligases.RNA. 2014 Apr;20(4):462-73. doi: 10.1261/rna.043752.113. Epub 2014 Feb 19. RNA. 2014. PMID: 24554441 Free PMC article.
-
Characterization of 3'-Phosphate RNA Ligase Paralogs RtcB1, RtcB2, and RtcB3 from Myxococcus xanthus Highlights DNA and RNA 5'-Phosphate Capping Activity of RtcB3.J Bacteriol. 2015 Nov;197(22):3616-24. doi: 10.1128/JB.00631-15. Epub 2015 Sep 8. J Bacteriol. 2015. PMID: 26350128 Free PMC article.
-
Structures of the noncanonical RNA ligase RtcB reveal the mechanism of histidine guanylylation.Biochemistry. 2013 Apr 16;52(15):2518-25. doi: 10.1021/bi4002375. Epub 2013 Apr 5. Biochemistry. 2013. PMID: 23560983 Free PMC article.
References
-
- Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. - PubMed
-
- Phizicky EM, Schwartz RC, Abelson J. Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J Biol Chem. 1986;261:2978–2986. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

