De novo automated design of small RNA circuits for engineering synthetic riboregulation in living cells
- PMID: 22949707
- PMCID: PMC3458397
- DOI: 10.1073/pnas.1203831109
De novo automated design of small RNA circuits for engineering synthetic riboregulation in living cells
Abstract
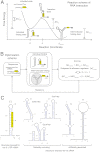
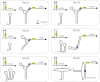
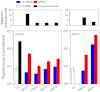
A grand challenge in synthetic biology is to use our current knowledge of RNA science to perform the automatic engineering of completely synthetic sequences encoding functional RNAs in living cells. We report here a fully automated design methodology and experimental validation of synthetic RNA interaction circuits working in a cellular environment. The computational algorithm, based on a physicochemical model, produces novel RNA sequences by exploring the space of possible sequences compatible with predefined structures. We tested our methodology in Escherichia coli by designing several positive riboregulators with diverse structures and interaction models, suggesting that only the energy of formation and the activation energy (free energy barrier to overcome for initiating the hybridization reaction) are sufficient criteria to engineer RNA interaction and regulation in bacteria. The designed sequences exhibit nonsignificant similarity to any known noncoding RNA sequence. Our riboregulatory devices work independently and in combination with transcription regulation to create complex logic circuits. Our results demonstrate that a computational methodology based on first-principles can be used to engineer interacting RNAs with allosteric behavior in living cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat Biotechnol. 2006;24:545–554. - PubMed
-
- Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr Opin Microbiol. 2007;10:102–109. - PubMed
-
- Rajkowitsch L, et al. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. - PubMed
-
- Pfleger BF, et al. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol. 2006;24:1027–1032. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

