XatA, an AT-1 autotransporter important for the virulence of Xylella fastidiosa Temecula1
- PMID: 22950010
- PMCID: PMC3426408
- DOI: 10.1002/mbo3.6
XatA, an AT-1 autotransporter important for the virulence of Xylella fastidiosa Temecula1
Abstract
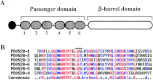
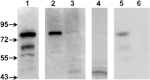
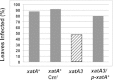
Xylella fastidiosa Temecula1 is the causative agent of Pierce's disease of grapevine, which is spread by xylem-feeding insects. An important feature of the infection cycle is the ability of X. fastidiosa to colonize and interact with two distinct environments, the xylem of susceptible plants and the insect foregut. Here, we describe our characterization of XatA, the X. fastidiosa autotransporter protein encoded by PD0528. XatA, which is classified as an AT-1 (classical) autotransporter, has a C-terminal β-barrel domain and a passenger domain composed of six tandem repeats of approximately 50 amino acids. Localization studies indicate that XatA is present in both the outer membrane and membrane vesicles and its passenger domain can be found in the supernatant. Moreover, XatA is important for X. fastidiosa autoaggregation and biofilm formation based on mutational analysis and the discovery that Escherichia coli expressing XatA acquire these traits. The xatA mutant also shows a significant decrease in Pierce's disease symptoms when inoculated into grapevines. Finally, X. fastidiosa homologs to XatA, which can be divided into three distinct groups based on synteny, form a single, well-supported clade, suggesting that they arose from a common ancestor.
Keywords: Adhesin; biofilm; sharpshooter; type V secretion; virulence factor.
Figures


Similar articles
-
Citrus and Coffee Strains of Xylella fastidiosa Induce Pierce's Disease in Grapevine.Plant Dis. 2002 Nov;86(11):1206-1210. doi: 10.1094/PDIS.2002.86.11.1206. Plant Dis. 2002. PMID: 30818468
-
Virulence Comparison of a Comprehensive Panel of Xylella fastidiosa Pierce's Disease Isolates from California.Plant Dis. 2024 Jun;108(6):1555-1564. doi: 10.1094/PDIS-09-23-1923-RE. Epub 2024 May 16. Plant Dis. 2024. PMID: 38105458
-
Infection of Blueberry Cultivar 'Emerald' with a California Pierce's Disease Strain of Xylella fastidiosa and Acquisition by Glassy-Winged Sharpshooter.Plant Dis. 2020 Jan;104(1):154-160. doi: 10.1094/PDIS-05-19-1126-RE. Epub 2019 Nov 6. Plant Dis. 2020. PMID: 31697223
-
Pierce's Disease of Grapevines: A Review of Control Strategies and an Outline of an Epidemiological Model.Front Microbiol. 2018 Sep 12;9:2141. doi: 10.3389/fmicb.2018.02141. eCollection 2018. Front Microbiol. 2018. PMID: 30258423 Free PMC article. Review.
-
The biology of xylem fluid-feeding insect vectors of Xylella fastidiosa and their relation to disease epidemiology.Annu Rev Entomol. 2004;49:243-70. doi: 10.1146/annurev.ento.49.061802.123403. Annu Rev Entomol. 2004. PMID: 14651464 Review.
Cited by
-
Potential complications when developing gene deletion clones in Xylella fastidiosa.BMC Res Notes. 2015 Apr 16;8:155. doi: 10.1186/s13104-015-1117-9. BMC Res Notes. 2015. PMID: 25880211 Free PMC article.
-
The Secreted Protease PrtA Controls Cell Growth, Biofilm Formation and Pathogenicity in Xylella fastidiosa.Sci Rep. 2016 Aug 5;6:31098. doi: 10.1038/srep31098. Sci Rep. 2016. PMID: 27492542 Free PMC article.
-
Characterization of the Xylella fastidiosa PD1311 gene mutant and its suppression of Pierce's disease on grapevines.Mol Plant Pathol. 2017 Jun;18(5):684-694. doi: 10.1111/mpp.12428. Epub 2016 Aug 22. Mol Plant Pathol. 2017. PMID: 27388152 Free PMC article.
-
The Type II Secreted Lipase/Esterase LesA is a Key Virulence Factor Required for Xylella fastidiosa Pathogenesis in Grapevines.Sci Rep. 2016 Jan 12;6:18598. doi: 10.1038/srep18598. Sci Rep. 2016. PMID: 26753904 Free PMC article.
-
Message in a Bubble: Shuttling Small RNAs and Proteins Between Cells and Interacting Organisms Using Extracellular Vesicles.Annu Rev Plant Biol. 2021 Jun 17;72:497-524. doi: 10.1146/annurev-arplant-081720-010616. Annu Rev Plant Biol. 2021. PMID: 34143650 Free PMC article. Review.
References
-
- Adindla S, Inampudi KK, Guruprasad L. Cell surface proteins in archaeal and bacterial genomes comprising “LVIVD”, “RIVW” and “LGxL” tandem sequence repeats are predicted to fold as β-propeller. Int. J. Biol. Macromol. 2007;41:454–468. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Biegert A, Soding J. De novo identification of highly diverged protein repeats by probabilistic consistency. Bioinformatics. 2008;24:807–814. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

