Modeling gene expression using chromatin features in various cellular contexts
- PMID: 22950368
- PMCID: PMC3491397
- DOI: 10.1186/gb-2012-13-9-r53
Modeling gene expression using chromatin features in various cellular contexts
Abstract
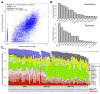
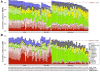
Background: Previous work has demonstrated that chromatin feature levels correlate with gene expression. The ENCODE project enables us to further explore this relationship using an unprecedented volume of data. Expression levels from more than 100,000 promoters were measured using a variety of high-throughput techniques applied to RNA extracted by different protocols from different cellular compartments of several human cell lines. ENCODE also generated the genome-wide mapping of eleven histone marks, one histone variant, and DNase I hypersensitivity sites in seven cell lines.
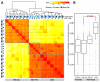
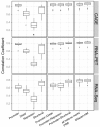
Results: We built a novel quantitative model to study the relationship between chromatin features and expression levels. Our study not only confirms that the general relationships found in previous studies hold across various cell lines, but also makes new suggestions about the relationship between chromatin features and gene expression levels. We found that expression status and expression levels can be predicted by different groups of chromatin features, both with high accuracy. We also found that expression levels measured by CAGE are better predicted than by RNA-PET or RNA-Seq, and different categories of chromatin features are the most predictive of expression for different RNA measurement methods. Additionally, PolyA+ RNA is overall more predictable than PolyA- RNA among different cell compartments, and PolyA+ cytosolic RNA measured with RNA-Seq is more predictable than PolyA+ nuclear RNA, while the opposite is true for PolyA- RNA.
Conclusions: Our study provides new insights into transcriptional regulation by analyzing chromatin features in different cellular contexts.
Figures


References
-
- Koch CM, Andrews RM, Flicek P, Dillon SC, Karaöz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Weng Z, Birney E, Carter NP, Vetrie D, Dunham I. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

