Characterization of copper-resistant bacteria and bacterial communities from copper-polluted agricultural soils of central Chile
- PMID: 22950448
- PMCID: PMC3496636
- DOI: 10.1186/1471-2180-12-193
Characterization of copper-resistant bacteria and bacterial communities from copper-polluted agricultural soils of central Chile
Abstract
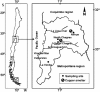
Background: Copper mining has led to Cu pollution in agricultural soils. In this report, the effects of Cu pollution on bacterial communities of agricultural soils from Valparaiso region, central Chile, were studied. Denaturing gradient gel electrophoresis (DGGE) of the 16S rRNA genes was used for the characterization of bacterial communities from Cu-polluted and non-polluted soils. Cu-resistant bacterial strains were isolated from Cu-polluted soils and characterized.
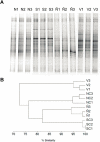
Results: DGGE showed a similar high number of bands and banding pattern of the bacterial communities from Cu-polluted and non-polluted soils. The presence of copA genes encoding the multi-copper oxidase that confers Cu-resistance in bacteria was detected by PCR in metagenomic DNA from the three Cu-polluted soils, but not in the non-polluted soil. The number of Cu-tolerant heterotrophic cultivable bacteria was significantly higher in Cu-polluted soils than in the non-polluted soil. Ninety two Cu-resistant bacterial strains were isolated from three Cu-polluted agricultural soils. Five isolated strains showed high resistance to copper (MIC ranged from 3.1 to 4.7 mM) and also resistance to other heavy metals. 16S rRNA gene sequence analyses indicate that these isolates belong to the genera Sphingomonas, Stenotrophomonas and Arthrobacter. The Sphingomonas sp. strains O12, A32 and A55 and Stenotrophomonas sp. C21 possess plasmids containing the Cu-resistance copA genes. Arthrobacter sp. O4 possesses the copA gene, but plasmids were not detected in this strain. The amino acid sequences of CopA from Sphingomonas isolates (O12, A32 and A55), Stenotrophomonas strain (C21) and Arthrobacter strain (O4) are closely related to CopA from Sphingomonas, Stenotrophomonas and Arthrobacter strains, respectively.
Conclusions: This study suggests that bacterial communities of agricultural soils from central Chile exposed to long-term Cu-pollution have been adapted by acquiring Cu genetic determinants. Five bacterial isolates showed high copper resistance and additional resistance to other heavy metals. Detection of copA gene in plasmids of four Cu-resistant isolates indicates that mobile genetic elements are involved in the spreading of Cu genetic determinants in polluted environments.
Figures
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

