Segmental dataset and whole body expression data do not support the hypothesis that non-random movement is an intrinsic property of Drosophila retrogenes
- PMID: 22950647
- PMCID: PMC3532075
- DOI: 10.1186/1471-2148-12-169
Segmental dataset and whole body expression data do not support the hypothesis that non-random movement is an intrinsic property of Drosophila retrogenes
Abstract
Background: Several studies in Drosophila have shown excessive movement of retrogenes from the X chromosome to autosomes, and that these genes are frequently expressed in the testis. This phenomenon has led to several hypotheses invoking natural selection as the process driving male-biased genes to the autosomes. Metta and Schlötterer (BMC Evol Biol 2010, 10:114) analyzed a set of retrogenes where the parental gene has been subsequently lost. They assumed that this class of retrogenes replaced the ancestral functions of the parental gene, and reported that these retrogenes, although mostly originating from movement out of the X chromosome, showed female-biased or unbiased expression. These observations led the authors to suggest that selective forces (such as meiotic sex chromosome inactivation and sexual antagonism) were not responsible for the observed pattern of retrogene movement out of the X chromosome.
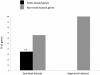
Results: We reanalyzed the dataset published by Metta and Schlötterer and found several issues that led us to a different conclusion. In particular, Metta and Schlötterer used a dataset combined with expression data in which significant sex-biased expression is not detectable. First, the authors used a segmental dataset where the genes selected for analysis were less testis-biased in expression than those that were excluded from the study. Second, sex-biased expression was defined by comparing male and female whole-body data and not the expression of these genes in gonadal tissues. This approach significantly reduces the probability of detecting sex-biased expressed genes, which explains why the vast majority of the genes analyzed (parental and retrogenes) were equally expressed in both males and females. Third, the female-biased expression observed by Metta and Schlötterer is mostly found for parental genes located on the X chromosome, which is known to be enriched with genes with female-biased expression. Fourth, using additional gonad expression data, we found that autosomal genes analyzed by Metta and Schlötterer are less up regulated in ovaries and have higher chance to be expressed in meiotic cells of spermatogenesis when compared to X-linked genes.
Conclusions: The criteria used to select retrogenes and the sex-biased expression data based on whole adult flies generated a segmental dataset of female-biased and unbiased expressed genes that was unable to detect the higher propensity of autosomal retrogenes to be expressed in males. Thus, there is no support for the authors' view that the movement of new retrogenes, which originated from X-linked parental genes, was not driven by selection. Therefore, selection-based genetic models remain the most parsimonious explanations for the observed chromosomal distribution of retrogenes.
Figures
Similar articles
-
Non-random genomic integration - an intrinsic property of retrogenes in Drosophila?BMC Evol Biol. 2010 Apr 28;10:114. doi: 10.1186/1471-2148-10-114. BMC Evol Biol. 2010. PMID: 20426838 Free PMC article.
-
Re-analysis of the larval testis data on meiotic sex chromosome inactivation revealed evidence for tissue-specific gene expression related to the drosophila X chromosome.BMC Biol. 2012 Jun 12;10:49; author reply 50. doi: 10.1186/1741-7007-10-49. BMC Biol. 2012. PMID: 22691264 Free PMC article.
-
General gene movement off the X chromosome in the Drosophila genus.Genome Res. 2009 May;19(5):897-903. doi: 10.1101/gr.088609.108. Epub 2009 Feb 27. Genome Res. 2009. PMID: 19251740 Free PMC article.
-
X chromosomes, retrogenes and their role in male reproduction.Trends Endocrinol Metab. 2004 Mar;15(2):79-83. doi: 10.1016/j.tem.2004.01.007. Trends Endocrinol Metab. 2004. PMID: 15036254 Review.
-
Gene content evolution on the X chromosome.Curr Opin Genet Dev. 2008 Dec;18(6):493-8. doi: 10.1016/j.gde.2008.09.006. Epub 2008 Oct 16. Curr Opin Genet Dev. 2008. PMID: 18929654 Free PMC article. Review.
Cited by
-
New gene evolution: little did we know.Annu Rev Genet. 2013;47:307-33. doi: 10.1146/annurev-genet-111212-133301. Epub 2013 Sep 13. Annu Rev Genet. 2013. PMID: 24050177 Free PMC article. Review.
References
-
- Dai H, Yoshimatsu TF, Long M. Retrogene movement within- and between-chromosomes in the evolution of Drosophila genomes. Gene. 2006;385:96–102. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

