Inferring genetic architecture of complex traits using Bayesian integrative analysis of genome and transcriptome data
- PMID: 22950759
- PMCID: PMC3543188
- DOI: 10.1186/1471-2164-13-456
Inferring genetic architecture of complex traits using Bayesian integrative analysis of genome and transcriptome data
Abstract
Background: To understand the genetic architecture of complex traits and bridge the genotype-phenotype gap, it is useful to study intermediate -omics data, e.g. the transcriptome. The present study introduces a method for simultaneous quantification of the contributions from single nucleotide polymorphisms (SNPs) and transcript abundances in explaining phenotypic variance, using Bayesian whole-omics models. Bayesian mixed models and variable selection models were used and, based on parameter samples from the model posterior distributions, explained variances were further partitioned at the level of chromosomes and genome segments.
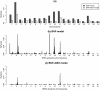
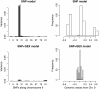
Results: We analyzed three growth-related traits: Body Weight (BW), Feed Intake (FI), and Feed Efficiency (FE), in an F2 population of 440 mice. The genomic variation was covered by 1806 tag SNPs, and transcript abundances were available from 23,698 probes measured in the liver. Explained variances were computed for models using pedigree, SNPs, transcripts, and combinations of these. Comparison of these models showed that for BW, a large part of the variation explained by SNPs could be covered by the liver transcript abundances; this was less true for FI and FE. For BW, the main quantitative trait loci (QTLs) are found on chromosomes 1, 2, 9, 10, and 11, and the QTLs on 1, 9, and 10 appear to be expression Quantitative Trait Locus (eQTLs) affecting gene expression in the liver. Chromosome 9 is the case of an apparent eQTL, showing that genomic variance disappears, and that a tri-modal distribution of genomic values collapses, when gene expressions are added to the model.
Conclusions: With increased availability of various -omics data, integrative approaches are promising tools for understanding the genetic architecture of complex traits. Partitioning of explained variances at the chromosome and genome-segment level clearly separated regulatory and structural genomic variation as the areas where SNP effects disappeared/remained after adding transcripts to the model. The models that include transcripts explained more phenotypic variance and were better at predicting phenotypes than a model using SNPs alone. The predictions from these Bayesian models are generally unbiased, validating the estimates of explained variances.
Figures

References
-
- Hayes B, Goddard ME. Break-even cost of genotyping genetic mutations affecting economic traits in Australian pig enterprises. Livest Prod Sci. 2004;89(2–3):235–242.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

