Metagenomics and metatranscriptomics: windows on CF-associated viral and microbial communities
- PMID: 22951208
- PMCID: PMC3534838
- DOI: 10.1016/j.jcf.2012.07.009
Metagenomics and metatranscriptomics: windows on CF-associated viral and microbial communities
Abstract
Background: Samples collected from CF patient airways often contain large amounts of host-derived nucleic acids that interfere with recovery and purification of microbial and viral nucleic acids. This study describes metagenomic and metatranscriptomic methods that address these issues.
Methods: Microbial and viral metagenomes, and microbial metatranscriptomes, were successfully prepared from sputum samples from five adult CF patients.
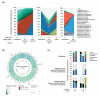
Results: Contaminating host DNA was dramatically reduced in the metagenomes. Each CF patient presented a unique microbiome; in some Pseudomonas aeruginosa was replaced by other opportunistic bacteria. Even though the taxonomic composition of the microbiomes is very different, the metabolic potentials encoded by the community are very similar. The viral communities were dominated by phages that infect major CF pathogens. The metatranscriptomes reveal differential expression of encoded metabolic potential with changing health status.
Conclusions: Microbial and viral metagenomics combined with microbial transcriptomics characterize the dynamic polymicrobial communities found in CF airways, revealing both the taxa present and their current metabolic activities. These approaches can facilitate the development of individualized treatment plans and novel therapeutic approaches.
Copyright © 2012 European Cystic Fibrosis Society. Published by Elsevier B.V. All rights reserved.
Figures



Similar articles
-
Biogeochemical forces shape the composition and physiology of polymicrobial communities in the cystic fibrosis lung.mBio. 2014 Mar 18;5(2):e00956-13. doi: 10.1128/mBio.00956-13. mBio. 2014. PMID: 24643867 Free PMC article.
-
Purifying the impure: sequencing metagenomes and metatranscriptomes from complex animal-associated samples.J Vis Exp. 2014 Dec 22;(94):52117. doi: 10.3791/52117. J Vis Exp. 2014. PMID: 25549184 Free PMC article.
-
Sputum DNA sequencing in cystic fibrosis: non-invasive access to the lung microbiome and to pathogen details.Microbiome. 2017 Feb 10;5(1):20. doi: 10.1186/s40168-017-0234-1. Microbiome. 2017. PMID: 28187782 Free PMC article.
-
The airway microbiome in cystic fibrosis and implications for treatment.Curr Opin Pediatr. 2011 Jun;23(3):319-24. doi: 10.1097/MOP.0b013e32834604f2. Curr Opin Pediatr. 2011. PMID: 21494150 Review.
-
The polymicrobial nature of airway infections in cystic fibrosis: Cangene Gold Medal Lecture.Can J Microbiol. 2011 Feb;57(2):69-77. doi: 10.1139/w10-105. Can J Microbiol. 2011. PMID: 21326348 Review.
Cited by
-
The cystic fibrosis lower airways microbial metagenome.ERJ Open Res. 2016 May 9;2(2):00096-2015. doi: 10.1183/23120541.00096-2015. eCollection 2016 Apr. ERJ Open Res. 2016. PMID: 27730195 Free PMC article.
-
Standardized multi-omics of Earth's microbiomes reveals microbial and metabolite diversity.Nat Microbiol. 2022 Dec;7(12):2128-2150. doi: 10.1038/s41564-022-01266-x. Epub 2022 Nov 28. Nat Microbiol. 2022. PMID: 36443458 Free PMC article.
-
Untargeted Metagenomic Investigation of the Airway Microbiome of Cystic Fibrosis Patients with Moderate-Severe Lung Disease.Microorganisms. 2020 Jul 4;8(7):1003. doi: 10.3390/microorganisms8071003. Microorganisms. 2020. PMID: 32635564 Free PMC article.
-
Pseudomonas aeruginosa Increases the Sensitivity of Biofilm-Grown Staphylococcus aureus to Membrane-Targeting Antiseptics and Antibiotics.mBio. 2019 Jul 30;10(4):e01501-19. doi: 10.1128/mBio.01501-19. mBio. 2019. PMID: 31363032 Free PMC article.
-
A restructuring of microbiome niche space is associated with Elexacaftor-Tezacaftor-Ivacaftor therapy in the cystic fibrosis lung.J Cyst Fibros. 2022 Nov;21(6):996-1005. doi: 10.1016/j.jcf.2021.11.003. Epub 2021 Nov 22. J Cyst Fibros. 2022. PMID: 34824018 Free PMC article.
References
-
- Boucher RC. An overview of the pathogenesis of Cystic Fibrosis lung disease. Advanced Drug Delivery Reviews. 2002 Dec 5;54(11):1359–71. - PubMed
-
- Riordan JR. CFTR function and prospects for therapy. Annu. Rev. Biochem. 2008;77:701–26. - PubMed
-
- Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Jones G, Bruce KD. Characterization of bacterial community diversity in Cystic Fibrosis lung infections by use of 16S ribosomal DNA terminal restriction fragment length polymorphism profiling. J. Clin. Microbiol. 2004 Nov 1;42(11):5176–83. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

