Do MHCII-presented neoantigens drive type 1 diabetes and other autoimmune diseases?
- PMID: 22951444
- PMCID: PMC3426820
- DOI: 10.1101/cshperspect.a007765
Do MHCII-presented neoantigens drive type 1 diabetes and other autoimmune diseases?
Abstract
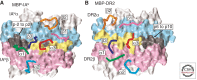
The strong association between particular MHCII alleles and type 1 diabetes is not fully understood. Two ideas that have been considered for many years are that autoimmunity is driven by (1) low-affinity CD4(+) T cells that escape thymic negative selection and respond to certain autoantigen peptides that are particularly well presented by particular MHCII molecules, or (2) CD4(+) T cells responding to neoantigens that are absent in the thymus, but uniquely created in the target tissue in the periphery and presented by particular MHCII alleles. Here we discuss the recent structural data in favor of the second idea. We review studies suggesting that peptide antigens recognized by autoimmune T cells are uniquely proteolytically processed and/or posttranslationally modified in the target tissue, thus allowing these T cells to escape deletion in the thymus during T-cell development. We postulate that an encounter with these tissue-specific neoantigenic peptides presented by the particular susceptible MHCII alleles in the peripheral tissues when accompanied by the appropriate inflammatory milieu activates these T-cell escapees leading to the onset of autoimmune disease.
Figures




References
-
- Abadie V, Sollid LM, Barreiro LB, Jabri B 2011. Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu Rev Immunol 29: 493–525 - PubMed
-
- Abiru N, Wegmann D, Kawasaki E, Gottlieb P, Simone E, Eisenbarth GS 2000. Dual overlapping peptides recognized by insulin peptide B:9-23 T cell receptor AV13S3 T cell clones of the NOD mouse. J Autoimmun 14: 231–237 - PubMed
-
- Anderton SM 2004. Post-translational modifications of self antigens: Implications for autoimmunity. Curr Opin Immunol 16: 753–758 - PubMed
-
- Bankovich AJ, Girvin AT, Moesta AK, Garcia KC 2004. Peptide register shifting within the MHC groove: Theory becomes reality. Mol Immunol 40: 1033–1039 - PubMed
-
- Burton AR, Vincent E, Arnold PY, Lennon GP, Smeltzer M, Li CS, Haskins K, Hutton J, Tisch RM, Sercarz EE, et al. 2008. On the pathogenicity of autoantigen-specific T-cell receptors. Diabetes 57: 1321–1330 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
