Structure of Crimean-Congo hemorrhagic fever virus nucleoprotein: superhelical homo-oligomers and the role of caspase-3 cleavage
- PMID: 22951837
- PMCID: PMC3486442
- DOI: 10.1128/JVI.01627-12
Structure of Crimean-Congo hemorrhagic fever virus nucleoprotein: superhelical homo-oligomers and the role of caspase-3 cleavage
Abstract
Crimean-Congo hemorrhagic fever, a severe hemorrhagic disease found throughout Africa, Europe, and Asia, is caused by the tick-borne Crimean-Congo hemorrhagic fever virus (CCHFV). CCHFV is a negative-sense single-stranded RNA (ssRNA) virus belonging to the Nairovirus genus of the Bunyaviridae family. Its genome of three single-stranded RNA segments is encapsidated by the nucleocapsid protein (CCHFV N) to form the ribonucleoprotein complex. This ribonucleoprotein complex is required during replication and transcription of the viral genomic RNA. Here, we present the crystal structures of the CCHFV N in two distinct forms, an oligomeric form comprised of double antiparallel superhelices and a monomeric form. The head-to-tail interaction of the stalk region of one CCHFV N subunit with the base of the globular body of the adjacent subunit stabilizes the helical organization of the oligomeric form of CCHFV N. It also masks the conserved caspase-3 cleavage site present at the tip of the stalk region from host cell caspase-3 interaction and cleavage. By incubation with primer-length ssRNAs, we also obtained the crystal structure of CCHFV N in its monomeric form, which is similar to a recently published structure. The conformational change of CCHFV N upon deoligomerization results in the exposure of the caspase-3 cleavage site and subjects CCHFV N to caspase-3 cleavage. Mutations of this cleavage site inhibit cleavage by caspase-3 and result in enhanced viral polymerase activity. Thus, cleavage of CCHFV N by host cell caspase-3 appears to be crucial for controlling viral RNA synthesis and represents an important host defense mechanism against CCHFV infection.
Figures
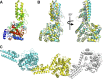
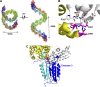

Similar articles
-
Crimean-Congo hemorrhagic fever virus nucleocapsid protein harbors distinct RNA-binding sites in the stalk and head domains.J Biol Chem. 2019 Mar 29;294(13):5023-5037. doi: 10.1074/jbc.RA118.004976. Epub 2019 Feb 5. J Biol Chem. 2019. PMID: 30723154 Free PMC article.
-
Molecular basis for the formation of ribonucleoprotein complex of Crimean-Congo hemorrhagic fever virus.J Struct Biol. 2016 Dec;196(3):455-465. doi: 10.1016/j.jsb.2016.09.013. Epub 2016 Sep 22. J Struct Biol. 2016. PMID: 27666016
-
Heat Shock Protein 70 Family Members Interact with Crimean-Congo Hemorrhagic Fever Virus and Hazara Virus Nucleocapsid Proteins and Perform a Functional Role in the Nairovirus Replication Cycle.J Virol. 2016 Sep 29;90(20):9305-16. doi: 10.1128/JVI.00661-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27512070 Free PMC article.
-
Crimean-Congo Hemorrhagic Fever.Lab Med. 2015 Summer;46(3):180-9. doi: 10.1309/LMN1P2FRZ7BKZSCO. Lab Med. 2015. PMID: 26199256 Review.
-
Epidemiological Aspects of Crimean-Congo Hemorrhagic Fever in Western Europe: What about the Future?Microorganisms. 2021 Mar 21;9(3):649. doi: 10.3390/microorganisms9030649. Microorganisms. 2021. PMID: 33801015 Free PMC article. Review.
Cited by
-
Crimean-Congo hemorrhagic fever virus nucleocapsid protein harbors distinct RNA-binding sites in the stalk and head domains.J Biol Chem. 2019 Mar 29;294(13):5023-5037. doi: 10.1074/jbc.RA118.004976. Epub 2019 Feb 5. J Biol Chem. 2019. PMID: 30723154 Free PMC article.
-
Crimean Congo Hemorrhagic Fever Virus for Clinicians-Virology, Pathogenesis, and Pathology.Emerg Infect Dis. 2024 May;30(5):847-853. doi: 10.3201/eid3005.231646. Emerg Infect Dis. 2024. PMID: 38666566 Free PMC article. Review.
-
Genomic Characterization of the Genus Nairovirus (Family Bunyaviridae).Viruses. 2016 Jun 10;8(6):164. doi: 10.3390/v8060164. Viruses. 2016. PMID: 27294949 Free PMC article.
-
Early Bunyavirus-Host Cell Interactions.Viruses. 2016 May 24;8(5):143. doi: 10.3390/v8050143. Viruses. 2016. PMID: 27213430 Free PMC article. Review.
-
Genomic Characterisation of Vinegar Hill Virus, An Australian Nairovirus Isolated in 1983 from Argas Robertsi Ticks Collected from Cattle Egrets.Viruses. 2017 Dec 5;9(12):373. doi: 10.3390/v9120373. Viruses. 2017. PMID: 29206186 Free PMC article.
References
-
- Albertini AA, et al. 2006. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313:360–363 - PubMed
-
- Andersson I, et al. 2004. Role of actin filaments in targeting of Crimean Congo hemorrhagic fever virus nucleocapsid protein to perinuclear regions of mammalian cells. J. Med. Virol. 72:83–93 - PubMed
-
- Brunger AT, et al. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

