Nucleic acid aptamers: an emerging frontier in cancer therapy
- PMID: 22951893
- PMCID: PMC3869973
- DOI: 10.1039/c2cc35042d
Nucleic acid aptamers: an emerging frontier in cancer therapy
Abstract
The last two decades have witnessed the development and application of nucleic acid aptamers in a variety of fields, including target analysis, disease therapy, and molecular and cellular engineering. The efficient and widely applicable aptamer selection, reproducible chemical synthesis and modification, generally impressive target binding selectivity and affinity, relatively rapid tissue penetration, low immunogenicity, and rapid systemic clearance make aptamers ideal recognition elements for use as therapeutics or for in vivo delivery of therapeutics. In this feature article, we discuss the development and biomedical application of nucleic acid aptamers, with emphasis on cancer cell aptamer isolation, targeted cancer therapy, oncology biomarker identification and drug discovery.
Figures




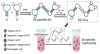
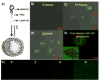
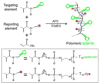
References
-
- Ellington AD, Szostak JW. Nature. 1990;346:818–822. - PubMed
-
- Tuerk C, Gold L. Science. 1990;249:505–510. - PubMed
-
- Huizenga DE, Szostak JW. Biochemistry. 1995;34:656–665. - PubMed
-
- Hermann T, Patel DJ. Science. 2000;287:820–825. - PubMed
-
- Mallikaratchy P, Stahelin RV, Cao Z, Cho W, Tan W. Chem Commun. 2006:3229–3231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

