SINE retrotransposons cause epigenetic reprogramming of adjacent gene promoters
- PMID: 22952045
- PMCID: PMC3475755
- DOI: 10.1158/1541-7786.MCR-12-0351
SINE retrotransposons cause epigenetic reprogramming of adjacent gene promoters
Abstract
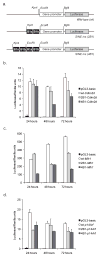
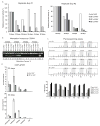
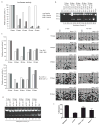
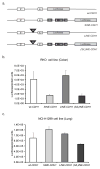
Almost half of the human genome and as much as 40% of the mouse genome is composed of repetitive DNA sequences. The majority of these repeats are retrotransposons of the SINE and LINE families, and such repeats are generally repressed by epigenetic mechanisms. It has been proposed that these elements can act as methylation centers from which DNA methylation spreads into gene promoters in cancer. Contradictory to a methylation center function, we have found that retrotransposons are enriched near promoter CpG islands that stay methylation-free in cancer. Clearly, it is important to determine which influence, if any, these repetitive elements have on nearby gene promoters. Using an in vitro system, we confirm here that SINE B1 elements can influence the activity of downstream gene promoters, with acquisition of DNA methylation and loss of activating histone marks, thus resulting in a repressed state. SINE sequences themselves did not immediately acquire DNA methylation but were marked by H3K9me2 and H3K27me3. Moreover, our bisulfite sequencing data did not support that gain of DNA methylation in gene promoters occurred by methylation spreading from SINE B1 repeats. Genome-wide analysis of SINE repeats distribution showed that their enrichment is directly correlated with the presence of USF1, USF2, and CTCF binding, proteins with insulator function. In summary, our work supports the concept that SINE repeats interfere negatively with gene expression and that their presence near gene promoters is counter-selected, except when the promoter is protected by an insulator element.
Conflict of interest statement
Figures



References
-
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–32. - PubMed
-
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–71. - PubMed
-
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

