Identification and quantification of a new family of peptide endocannabinoids (Pepcans) showing negative allosteric modulation at CB1 receptors
- PMID: 22952224
- PMCID: PMC3481297
- DOI: 10.1074/jbc.M112.382481
Identification and quantification of a new family of peptide endocannabinoids (Pepcans) showing negative allosteric modulation at CB1 receptors
Abstract
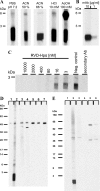
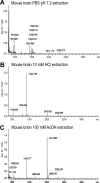
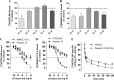
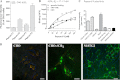
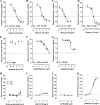
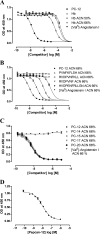
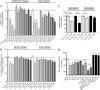
The α-hemoglobin-derived dodecapeptide RVD-hemopressin (RVDPVNFKLLSH) has been proposed to be an endogenous agonist for the cannabinoid receptor type 1 (CB(1)). To study this peptide, we have raised mAbs against its C-terminal part. Using an immunoaffinity mass spectrometry approach, a whole family of N-terminally extended peptides in addition to RVD-Hpα were identified in rodent brain extracts and human and mouse plasma. We designated these peptides Pepcan-12 (RVDPVNFKLLSH) to Pepcan-23 (SALSDLHAHKLRVDPVNFKLLSH), referring to peptide length. The most abundant Pepcans found in the brain were tested for CB(1) receptor binding. In the classical radioligand displacement assay, Pepcan-12 was the most efficacious ligand but only partially displaced both [(3)H]CP55,940 and [(3)H]WIN55,212-2. The data were fitted with the allosteric ternary complex model, revealing a cooperativity factor value α < 1, thus indicating a negative allosteric modulation. Dissociation kinetic studies of [(3)H]CP55,940 in the absence and presence of Pepcan-12 confirmed these results by showing increased dissociation rate constants induced by Pepcan-12. A fluorescently labeled Pepcan-12 analog was synthesized to investigate the binding to CB(1) receptors. Competition binding studies revealed K(i) values of several Pepcans in the nanomolar range. Accordingly, using competitive ELISA, we found low nanomolar concentrations of Pepcans in human plasma and ∼100 pmol/g in mouse brain. Surprisingly, Pepcan-12 exhibited potent negative allosteric modulation of the orthosteric agonist-induced cAMP accumulation, [(35)S]GTPγS binding, and CB(1) receptor internalization. Pepcans are the first endogenous allosteric modulators identified for CB(1) receptors. Given their abundance in the brain, Pepcans could play an important physiological role in modulating endocannabinoid signaling.
Figures



References
-
- Matsuda L. A., Lolait S. J., Brownstein M. J., Young A. C., Bonner T. I. (1990) Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature 346, 561–564 - PubMed
-
- Munro S., Thomas K. L., Abu-Shaar M. (1993) Molecular characterization of a peripheral receptor for cannabinoids. Nature 365, 61–65 - PubMed
-
- Pertwee R. G., Howlett A. C., Abood M. E., Alexander S. P., Di Marzo V., Elphick M. R., Greasley P. J., Hansen H. S., Kunos G., Mackie K., Mechoulam R., Ross R. A. (2010) International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid receptors and their ligands. Beyond CB1 and CB2. Pharmacol. Rev. 62, 588–631 - PMC - PubMed
-
- Mackie K. (2006) Cannabinoid receptors as therapeutic targets. Annu. Rev. Pharmacol. Toxicol. 46, 101–122 - PubMed
-
- Hanus L. O. (2009) Pharmacological and therapeutic secrets of plant and brain (endo)cannabinoids. Med. Res. Rev. 29, 213–271 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

