T-cell receptor-optimized peptide skewing of the T-cell repertoire can enhance antigen targeting
- PMID: 22952231
- PMCID: PMC3481325
- DOI: 10.1074/jbc.M112.386409
T-cell receptor-optimized peptide skewing of the T-cell repertoire can enhance antigen targeting
Abstract
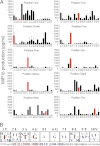
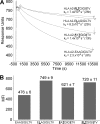
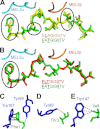
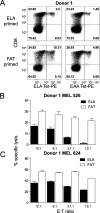
Altered peptide antigens that enhance T-cell immunogenicity have been used to improve peptide-based vaccination for a range of diseases. Although this strategy can prime T-cell responses of greater magnitude, the efficacy of constituent T-cell clonotypes within the primed population can be poor. To overcome this limitation, we isolated a CD8(+) T-cell clone (MEL5) with an enhanced ability to recognize the HLA A*0201-Melan A(27-35) (HLA A*0201-AAGIGILTV) antigen expressed on the surface of malignant melanoma cells. We used combinatorial peptide library screening to design an optimal peptide sequence that enhanced functional activation of the MEL5 clone, but not other CD8(+) T-cell clones that recognized HLA A*0201-AAGIGILTV poorly. Structural analysis revealed the potential for new contacts between the MEL5 T-cell receptor and the optimized peptide. Furthermore, the optimized peptide was able to prime CD8(+) T-cell populations in peripheral blood mononuclear cell isolates from multiple HLA A*0201(+) individuals that were capable of efficient HLA A*0201(+) melanoma cell destruction. This proof-of-concept study demonstrates that it is possible to design altered peptide antigens for the selection of superior T-cell clonotypes with enhanced antigen recognition properties.
Figures



References
-
- Davis M. M., Bjorkman P. J. (1988) T-cell antigen receptor genes and T-cell recognition. Nature 334, 395–402 - PubMed
-
- Garcia K. C., Teyton L., Wilson I. A. (1999) Structural basis of T cell recognition. Annu. Rev. Immunol. 17, 369–397 - PubMed
-
- Arstila T. P., Casrouge A., Baron V., Even J., Kanellopoulos J., Kourilsky P. (1999) A direct estimate of the human αβ T cell receptor diversity. Science 286, 958–961 - PubMed
-
- Mason D. (1998) A very high level of cross-reactivity is an essential feature of the T-cell receptor. Immunol. Today 19, 395–404 - PubMed
-
- Wooldridge L., Ekeruche-Makinde J., van den Berg H. A., Skowera A., Miles J. J., Tan M. P., Dolton G., Clement M., Llewellyn-Lacey S., Price D. A., Peakman M., Sewell A. K. (2012) A single autoimmune T cell receptor recognizes more than a million different peptides. J. Biol. Chem. 287, 1168–1177 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

