Kinetic modelling of GlmU reactions - prioritization of reaction for therapeutic application
- PMID: 22952829
- PMCID: PMC3428340
- DOI: 10.1371/journal.pone.0043969
Kinetic modelling of GlmU reactions - prioritization of reaction for therapeutic application
Abstract
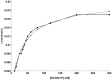
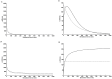
Mycobacterium tuberculosis(Mtu), a successful pathogen, has developed resistance against the existing anti-tubercular drugs necessitating discovery of drugs with novel action. Enzymes involved in peptidoglycan biosynthesis are attractive targets for antibacterial drug discovery. The bifunctional enzyme mycobacterial GlmU (Glucosamine 1-phosphate N-acetyltransferase/ N-acetylglucosamine-1-phosphate uridyltransferase) has been a target enzyme for drug discovery. Its C- and N- terminal domains catalyze acetyltransferase (rxn-1) and uridyltransferase (rxn-2) activities respectively and the final product is involved in peptidoglycan synthesis. However, the bifunctional nature of GlmU poses difficulty in deciding which function to be intervened for therapeutic advantage. Genetic analysis showed this as an essential gene but it is still unclear whether any one or both of the activities are critical for cell survival. Often enzymatic activity with suitable high-throughput assay is chosen for random screening, which may not be the appropriate biological function inhibited for maximal effect. Prediction of rate-limiting function by dynamic network analysis of reactions could be an option to identify the appropriate function. With a view to provide insights into biochemical assays with appropriate activity for inhibitor screening, kinetic modelling studies on GlmU were undertaken. Kinetic model of Mtu GlmU-catalyzed reactions was built based on the available kinetic data on Mtu and deduction from Escherichia coli data. Several model variants were constructed including coupled/decoupled, varying metabolite concentrations and presence/absence of product inhibitions. This study demonstrates that in coupled model at low metabolite concentrations, inhibition of either of the GlmU reactions cause significant decrement in the overall GlmU rate. However at higher metabolite concentrations, rxn-2 showed higher decrement. Moreover, with available intracellular concentration of the metabolites and in vivo variant of model, uncompetitive inhibition of rxn-2 caused highest decrement. Thus, at physiologically relevant metabolite concentrations, targeting uridyltranferase activity of Mtu GlmU would be a better choice for therapeutic intervention.
Conflict of interest statement
Figures

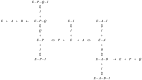


References
-
- Who (2010) World Health Organization: Tuberculosis Data.
-
- Zhou Y, Xin Y, Sha S, Ma Y (2011) Kinetic properties of Mycobacterium tuberculosis bifunctional GlmU. Archives of Microbiology 193: 751–757. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

