Allele-biased expression in differentiating human neurons: implications for neuropsychiatric disorders
- PMID: 22952857
- PMCID: PMC3431331
- DOI: 10.1371/journal.pone.0044017
Allele-biased expression in differentiating human neurons: implications for neuropsychiatric disorders
Abstract
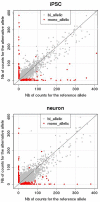
Stochastic processes and imprinting, along with genetic factors, lead to monoallelic or allele-biased gene expression. Stochastic monoallelic expression fine-tunes information processing in immune cells and the olfactory system, and imprinting plays an important role in development. Recent studies suggest that both stochastic events and imprinting may be more widespread than previously considered. We are interested in allele-biased gene expression occurring in the brain because parent-of-origin effects suggestive of imprinting appear to play a role in the transmission of schizophrenia (SZ) and autism spectrum disorders (ASD) in some families. In addition, allele-biased expression could help explain monozygotic (MZ) twin discordance and reduced penetrance. The ability to study allele-biased expression in human neurons has been transformed with the advent of induced pluripotent stem cell (iPSC) technology and next generation sequencing. Using transcriptome sequencing (RNA-Seq) we identified 801 genes in differentiating neurons that were expressed in an allele-biased manner. These included a number of putative SZ and ASD candidates, such as A2BP1 (RBFOX1), ERBB4, NLGN4X, NRG1, NRG3, NRXN1, and NLGN1. Overall, there was a modest enrichment for SZ and ASD candidate genes among those that showed evidence for allele-biased expression (chi-square, p = 0.02). In addition to helping explain MZ twin discordance and reduced penetrance, the capacity to group many candidate genes affecting a variety of molecular and cellular pathways under a common regulatory process - allele-biased expression - could have therapeutic implications.
Conflict of interest statement
Figures



References
-
- Delaval K, Feil R (2004) Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev 14(2) 188–195: 10.1016/j.gde.2004.01.005. - PubMed
-
- Holmes R, Soloway PD (2006) Regulation of imprinted DNA methylation. Cytogenet Genome Res 113(1–4) 122–129: 10.1159/000090823. - PubMed
-
- Radford EJ, Ferron SR, Ferguson-Smith AC (2011) Genomic imprinting as an adaptative model of developmental plasticity. FEBS Lett 585(13) 2059–2066: 10.1016/j.febslet.2011.05.063. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

