G4 motifs in human genes
- PMID: 22954217
- PMCID: PMC3806501
- DOI: 10.1111/j.1749-6632.2012.06586.x
G4 motifs in human genes
Abstract
The G4 motif, G(≥3) N(x) G(≥3) N(x) G(≥3) N(x) G(≥3) , is enriched in some genomic regions and depleted in others. This motif confers the ability to form an unusual four-stranded DNA structure, G4 DNA. G4 DNA is associated with genomic instability, which may explain depletion of G4 motifs from some genes and genomic regions. Conversely, G4 motifs are enriched downstream of transcription start sites, where they correlate with pausing. The uneven distribution of G4 motifs in the genome strongly suggests that mechanisms of selection act not only on one-dimensional genomic sequence, but also on structures formed by genomic DNA. The biological roles of G4 structures illustrate that, to understand genome function, it is important to consider the dynamic structural potential implicit in the G4 motif.
© 2012 New York Academy of Sciences.
Conflict of interest statement
The author declares no conflicts of interest.
Figures
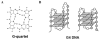


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

