The population genetics of cooperative gene regulation
- PMID: 22954408
- PMCID: PMC3537746
- DOI: 10.1186/1471-2148-12-173
The population genetics of cooperative gene regulation
Abstract
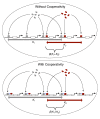
Background: Changes in gene regulatory networks drive the evolution of phenotypic diversity both within and between species. Rewiring of transcriptional networks is achieved either by changes to transcription factor binding sites or by changes to the physical interactions among transcription factor proteins. It has been suggested that the evolution of cooperative binding among factors can facilitate the adaptive rewiring of a regulatory network.
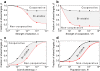
Results: We use a population-genetic model to explore when cooperative binding of transcription factors is favored by evolution, and what effects cooperativity then has on the adaptive re-writing of regulatory networks. We consider a pair of transcription factors that regulate multiple targets and overlap in the sets of target genes they regulate. We show that, under stabilising selection, cooperative binding between the transcription factors is favoured provided the amount of overlap between their target genes exceeds a threshold. The value of this threshold depends on several population-genetic factors: strength of selection on binding sites, cost of pleiotropy associated with protein-protein interactions, rates of mutation and population size. Once it is established, we find that cooperative binding of transcription factors significantly accelerates the adaptive rewiring of transcriptional networks under positive selection. We compare our qualitative predictions to systematic data on Saccharomyces cerevisiae transcription factors, their binding sites, and their protein-protein interactions.
Conclusions: Our study reveals a rich set of evolutionary dynamics driven by a tradeoff between the beneficial effects of cooperative binding at targets shared by a pair of factors, and the detrimental effects of cooperative binding for non-shared targets. We find that cooperative regulation will evolve when transcription factors share a sufficient proportion of their target genes. These findings help to explain empirical pattens in datasets of transcription factors in Saccharomyces cerevisiae and, they suggest that changes to physical interactions between transcription factors can play a critical role in the evolution of gene regulatory networks.
Figures


Similar articles
-
Evolution of transcriptional networks in yeast: alternative teams of transcriptional factors for different species.BMC Genomics. 2016 Nov 11;17(Suppl 10):826. doi: 10.1186/s12864-016-3102-7. BMC Genomics. 2016. PMID: 28185554 Free PMC article.
-
Adaptive evolution of transcription factor binding sites.BMC Evol Biol. 2004 Oct 28;4:42. doi: 10.1186/1471-2148-4-42. BMC Evol Biol. 2004. PMID: 15511291 Free PMC article.
-
The evolution of genetic networks by non-adaptive processes.Nat Rev Genet. 2007 Oct;8(10):803-13. doi: 10.1038/nrg2192. Nat Rev Genet. 2007. PMID: 17878896 Review.
-
The evolution of complex gene regulation by low-specificity binding sites.Proc Biol Sci. 2013 Aug 14;280(1768):20131313. doi: 10.1098/rspb.2013.1313. Print 2013 Oct 7. Proc Biol Sci. 2013. PMID: 23945682 Free PMC article.
-
Transcriptional regulation and the evolution of development.Int J Dev Biol. 2003;47(7-8):675-84. Int J Dev Biol. 2003. PMID: 14756343 Review.
Cited by
-
The rewiring of transcription circuits in evolution.Curr Opin Genet Dev. 2017 Dec;47:121-127. doi: 10.1016/j.gde.2017.09.004. Epub 2017 Nov 8. Curr Opin Genet Dev. 2017. PMID: 29120735 Free PMC article. Review.
-
Simulations of enhancer evolution provide mechanistic insights into gene regulation.Mol Biol Evol. 2014 Jan;31(1):184-200. doi: 10.1093/molbev/mst170. Epub 2013 Oct 4. Mol Biol Evol. 2014. PMID: 24097306 Free PMC article.
-
Inference of time-delayed gene regulatory networks based on dynamic Bayesian network hybrid learning method.Oncotarget. 2017 Sep 23;8(46):80373-80392. doi: 10.18632/oncotarget.21268. eCollection 2017 Oct 6. Oncotarget. 2017. PMID: 29113310 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

