A label-free proteome analysis strategy for identifying quantitative changes in erythrocyte membranes induced by red cell disorders
- PMID: 22954596
- PMCID: PMC3508302
- DOI: 10.1016/j.jprot.2012.08.010
A label-free proteome analysis strategy for identifying quantitative changes in erythrocyte membranes induced by red cell disorders
Abstract
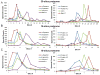
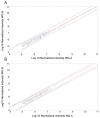
Red blood cells have been extensively studied but many questions regarding membrane properties and pathophysiology remain unanswered. Proteome analysis of red cell membranes is complicated by a very wide dynamic range of protein concentrations as well as the presence of proteins that are very large, very hydrophobic, or heterogeneously glycosylated. This study investigated the removal of other blood cell types, red cell membrane extraction, differing degrees of fractionation using 1-D SDS gels, and label-free quantitative methods to determine optimized conditions for proteomic comparisons of clinical blood samples. The results showed that fractionation of red cell membranes on 1-D SDS gels was more efficient than low-ionic-strength extractions followed by 1-D gel fractionation. When gel lanes were sliced into 30 uniform slices, a good depth of analysis that included the identification of most well-characterized, low-abundance red cell membrane proteins including those present at 500 to 10,000 copies per cell was obtained. Furthermore, the size separation enabled detection of changes due to proteolysis or in vivo protein crosslinking. A combination of Rosetta Elucidator quantitation and subsequent statistical analysis enabled the robust detection of protein differences that could be used to address unresolved questions in red cell disorders. This article is part of a Special Issue entitled: Integrated omics.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures




References
-
- Bosman GJ, Lasonder E, Luten M, Roerdinkholder-Stoelwinder B, Novotny VM, Bos H, De Grip WJ. The proteome of red cell membranes and vesicles during storage in blood bank conditions. Transfusion. 2008;48:827–35. - PubMed
-
- Low TY, Seow TK, Chung MCM. Separation of human erythrocyte membrane associated proteins with one-dimensional and two-dimensional gel electrophoresis followed by identification with matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics. 2002;2:1229–39. - PubMed
-
- Kakhniashvili DG, Bulla LA, Goodman SR. The human erythrocyte proteome - Analysis by ion trap mass spectrometry. Molecular & Cellular Proteomics. 2004;3:501–9. - PubMed
-
- De Palma A, Roveri A, Zaccarin M, Benazzi L, Daminelli S, Pantano G, Buttarello M, Ursini F, Gion M, Mauri PL. Extraction methods of red blood cell membrane proteins for Multidimensional Protein Identification Technology (MudPIT) analysis. Journal of Chromatography A. 2010;1217:5328–36. - PubMed
-
- Peker S, Akar N, Demiralp DO. Proteomic identification of erythrocyte membrane protein deficiency in hereditary spherocytosis. Molecular Biology Reports. 2012;39:3161–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

