Organic carbon transformations in high-Arctic peat soils: key functions and microorganisms
- PMID: 22955232
- PMCID: PMC3554415
- DOI: 10.1038/ismej.2012.99
Organic carbon transformations in high-Arctic peat soils: key functions and microorganisms
Abstract
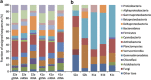
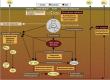
A substantial part of the Earths' soil organic carbon (SOC) is stored in Arctic permafrost peatlands, which represent large potential sources for increased emissions of the greenhouse gases CH(4) and CO(2) in a warming climate. The microbial communities and their genetic repertoire involved in the breakdown and mineralisation of SOC in these soils are, however, poorly understood. In this study, we applied a combined metagenomic and metatranscriptomic approach on two Arctic peat soils to investigate the identity and the gene pool of the microbiota driving the SOC degradation in the seasonally thawed active layers. A large and diverse set of genes encoding plant polymer-degrading enzymes was found, comparable to microbiotas from temperate and subtropical soils. This indicates that the metabolic potential for SOC degradation in Arctic peat is not different from that of other climatic zones. The majority of these genes were assigned to three bacterial phyla, Actinobacteria, Verrucomicrobia and Bacteroidetes. Anaerobic metabolic pathways and the fraction of methanogenic archaea increased with peat depth, evident for a gradual transition from aerobic to anaerobic lifestyles. A population of CH(4)-oxidising bacteria closely related to Methylobacter tundripaludum was the dominating active group of methanotrophs. Based on the in-depth characterisation of the microbes and their genes, we conclude that these Arctic peat soils will turn into CO(2) sources owing to increased active layer depth and prolonged growing season. However, the extent of future CH(4) emissions will critically depend on the response of the methanotrophic bacteria.
Figures


References
-
- Alves RJE.2011Ammonia-oxidizing archaea from High Arctic soilsMaster thesis, University of Lisbon: Lisbon, Portugal
-
- Berg MP, Bengtsson J. Temporal and spatial variability in soil food web structure. Oikos. 2007;116:1789–1804.
-
- Cao M, Marshall S, Gregson K. Global carbon exchange and methane emissions from natural wetlands: application of a process-based model. J Geophys Res. 1996;101:399–414.

