ATF4-dependent regulation of the JMJD3 gene during amino acid deprivation can be rescued in Atf4-deficient cells by inhibition of deacetylation
- PMID: 22955275
- PMCID: PMC3476305
- DOI: 10.1074/jbc.M112.399600
ATF4-dependent regulation of the JMJD3 gene during amino acid deprivation can be rescued in Atf4-deficient cells by inhibition of deacetylation
Abstract
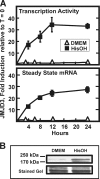
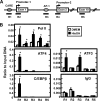
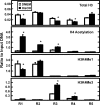
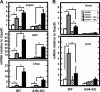
Following amino acid deprivation, the amino acid response (AAR) induces transcription from specific genes through a collection of signaling mechanisms, including the GCN2-eIF2-ATF4 pathway. The present report documents that the histone demethylase JMJD3 is an activating transcription factor 4 (ATF4)-dependent target gene. The JMJD3 gene contains two AAR-induced promoter activities and chromatin immunoprecipitation (ChIP) analysis showed that the AAR leads to enhanced ATF4 recruitment to the C/EBP-ATF response element (CARE) upstream of Promoter-1. AAR-induced histone modifications across the JMJD3 gene locus occur upon ATF4 binding. Jmjd3 transcription is not induced in Atf4-knock-out cells, but the AAR-dependent activation was rescued by inhibition of histone deacetylation with trichostatin A (TSA). The TSA rescue of AAR activation in the absence of Atf4 also occurred for the Atf3 and C/EBP homology protein (Chop) genes, but not for the asparagine synthetase gene. ChIP analysis of the Jmjd3, Atf3, and Chop genes in Atf4 knock-out cells documented that activation of the AAR in the presence of TSA led to specific changes in acetylation of histone H4. The results suggest that a primary function of ATF4 is to recruit histone acetyltransferase activity to a sub-set of AAR target genes. Thus, absolute binding of ATF4 to these particular genes is not required and no ATF4 interaction with the general transcription machinery is necessary. The data are consistent with the hypothesis that ATF4 functions as a pioneer factor to alter chromatin structure and thus, enhance transcription in a gene-specific manner.
Figures


Similar articles
-
Elevated cJUN expression and an ATF/CRE site within the ATF3 promoter contribute to activation of ATF3 transcription by the amino acid response.Physiol Genomics. 2013 Feb 15;45(4):127-37. doi: 10.1152/physiolgenomics.00160.2012. Epub 2012 Dec 26. Physiol Genomics. 2013. PMID: 23269699 Free PMC article.
-
Despite increased ATF4 binding at the C/EBP-ATF composite site following activation of the unfolded protein response, system A transporter 2 (SNAT2) transcription activity is repressed in HepG2 cells.J Biol Chem. 2008 Oct 10;283(41):27736-27747. doi: 10.1074/jbc.M803781200. Epub 2008 Aug 12. J Biol Chem. 2008. PMID: 18697751 Free PMC article.
-
ATF2 is required for amino acid-regulated transcription by orchestrating specific histone acetylation.Nucleic Acids Res. 2007;35(4):1312-21. doi: 10.1093/nar/gkm038. Epub 2007 Jan 31. Nucleic Acids Res. 2007. PMID: 17267404 Free PMC article.
-
ATF4-dependent transcription mediates signaling of amino acid limitation.Trends Endocrinol Metab. 2009 Nov;20(9):436-43. doi: 10.1016/j.tem.2009.05.008. Epub 2009 Sep 30. Trends Endocrinol Metab. 2009. PMID: 19800252 Free PMC article. Review.
-
Histone-modifying enzymes: regulators of developmental decisions and drivers of human disease.Epigenomics. 2012 Apr;4(2):163-77. doi: 10.2217/epi.12.3. Epigenomics. 2012. PMID: 22449188 Free PMC article. Review.
Cited by
-
Tumor suppressor BTG1 promotes PRMT1-mediated ATF4 function in response to cellular stress.Oncotarget. 2016 Jan 19;7(3):3128-43. doi: 10.18632/oncotarget.6519. Oncotarget. 2016. PMID: 26657730 Free PMC article.
-
Targeting Asparagine Metabolism in Solid Tumors.Nutrients. 2025 Jan 3;17(1):179. doi: 10.3390/nu17010179. Nutrients. 2025. PMID: 39796613 Free PMC article. Review.
-
Asparagine synthetase: regulation by cell stress and involvement in tumor biology.Am J Physiol Endocrinol Metab. 2013 Apr 15;304(8):E789-99. doi: 10.1152/ajpendo.00015.2013. Epub 2013 Feb 12. Am J Physiol Endocrinol Metab. 2013. PMID: 23403946 Free PMC article. Review.
-
Influence of Amino Acid Metabolism on Embryonic Stem Cell Function and Differentiation.Adv Nutr. 2016 Jul 15;7(4):780S-9S. doi: 10.3945/an.115.011031. Print 2016 Jul. Adv Nutr. 2016. PMID: 27422515 Free PMC article. Review.
-
DNA methylation entropy is associated with DNA sequence features and developmental epigenetic divergence.Nucleic Acids Res. 2023 Mar 21;51(5):2046-2065. doi: 10.1093/nar/gkad050. Nucleic Acids Res. 2023. PMID: 36762477 Free PMC article.
References
-
- Lillycrop K. A., Phillips E. S., Jackson A. A., Hanson M. A., Burdge G. C. (2005) Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J. Nutr. 135, 1382–1386 - PubMed
-
- Burdge G. C., Slater-Jefferies J., Torrens C., Phillips E. S., Hanson M. A., Lillycrop K. A. (2007) Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. Br. J. Nutr. 97, 435–439 - PMC - PubMed
-
- Zheng S., Rollet M., Pan Y. X. (2011) Maternal protein restriction during pregnancy induces CCAAT/enhancer-binding protein (C/EBPβ) expression through the regulation of histone modification at its promoter region in female offspring rat skeletal muscle. Epigenetics 6, 161–170 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

