Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia
- PMID: 22955920
- PMCID: PMC3482861
- DOI: 10.1182/blood-2012-03-415448
Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia
Abstract
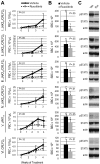
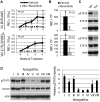
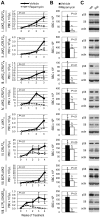
CRLF2 rearrangements, JAK1/2 point mutations, and JAK2 fusion genes have been identified in Philadelphia chromosome (Ph)-like acute lymphoblastic leukemia (ALL), a recently described subtype of pediatric high-risk B-precursor ALL (B-ALL) which exhibits a gene expression profile similar to Ph-positive ALL and has a poor prognosis. Hyperactive JAK/STAT and PI3K/mammalian target of rapamycin (mTOR) signaling is common in this high-risk subset. We, therefore, investigated the efficacy of the JAK inhibitor ruxolitinib and the mTOR inhibitor rapamycin in xenograft models of 8 pediatric B-ALL cases with and without CRLF2 and JAK genomic lesions. Ruxolitinib treatment yielded significantly lower peripheral blast counts compared with vehicle (P < .05) in 6 of 8 human leukemia xenografts and lower splenic blast counts (P < .05) in 8 of 8 samples. Enhanced responses to ruxolitinib were observed in samples harboring JAK-activating lesions and higher levels of STAT5 phosphorylation. Rapamycin controlled leukemia burden in all 8 B-ALL samples. Survival analysis of 2 representative B-ALL xenografts demonstrated prolonged survival with rapamycin treatment compared with vehicle (P < .01). These data demonstrate preclinical in vivo efficacy of ruxolitinib and rapamycin in this high-risk B-ALL subtype, for which novel treatments are urgently needed, and highlight the therapeutic potential of targeted kinase inhibition in Ph-like ALL.
Figures


References
-
- Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K12CA076931/CA/NCI NIH HHS/United States
- K12 CA076931/CA/NCI NIH HHS/United States
- R01 CA102646/CA/NCI NIH HHS/United States
- U01 CA157937/CA/NCI NIH HHS/United States
- U10 CA098543/CA/NCI NIH HHS/United States
- T32 CA009615/CA/NCI NIH HHS/United States
- T32CA009615-21/CA/NCI NIH HHS/United States
- CA102646/CA/NCI NIH HHS/United States
- T32 CA128583/CA/NCI NIH HHS/United States
- CA1116660/CA/NCI NIH HHS/United States
- U10 CA98413/CA/NCI NIH HHS/United States
- CA98543/CA/NCI NIH HHS/United States
- L40 CA142226/CA/NCI NIH HHS/United States
- U10 CA098413/CA/NCI NIH HHS/United States
- U24 CA114766/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

