IL-7-producing stromal cells are critical for lymph node remodeling
- PMID: 22955921
- PMCID: PMC3952724
- DOI: 10.1182/blood-2012-03-416859
IL-7-producing stromal cells are critical for lymph node remodeling
Abstract
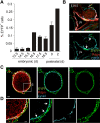
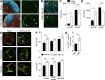
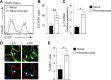
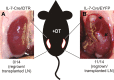
Nonhematopoietic stromal cells of secondary lymphoid organs form important scaffold and fluid transport structures, such as lymph node (LN) trabeculae, lymph vessels, and conduits. Furthermore, through the production of chemokines and cytokines, these cells generate a particular microenvironment that determines lymphocyte positioning and supports lymphocyte homeostasis. IL-7 is an important stromal cell-derived cytokine that has been considered to be derived mainly from T-cell zone fibroblastic reticular cells. We show here that lymphatic endothelial cells (LECs) are a prominent source of IL-7 both in human and murine LNs. Using bacterial artificial chromosome transgenic IL-7-Cre mice, we found that fibroblastic reticular cells and LECs strongly up-regulated IL-7 expression during LN remodeling after viral infection and LN reconstruction after avascular transplantation. Furthermore, IL-7-producing stromal cells contributed to de novo formation of LyveI-positive lymphatic structures connecting reconstructed LNs with the surrounding tissue. Importantly, diphtheria toxin-mediated depletion of IL-7-producing stromal cells completely abolished LN reconstruction. Taken together, this study identifies LN LECs as a major source of IL-7 and shows that IL-7-producing stromal cells are critical for reconstruction and remodeling of the distinct LN microenvironment.
Figures



References
-
- Corcoran AE, Riddell A, Krooshoop D, Venkitaraman AR. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature. 1998;391(6670):904–907. - PubMed
-
- Bertolino E, Reddy K, Medina KL, et al. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT5. Nat Immunol. 2005;6(8):836–843. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

