Evidence of abundant purifying selection in humans for recently acquired regulatory functions
- PMID: 22956687
- PMCID: PMC4104271
- DOI: 10.1126/science.1225057
Evidence of abundant purifying selection in humans for recently acquired regulatory functions
Abstract
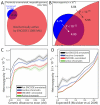
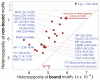
Although only 5% of the human genome is conserved across mammals, a substantially larger portion is biochemically active, raising the question of whether the additional elements evolve neutrally or confer a lineage-specific fitness advantage. To address this question, we integrate human variation information from the 1000 Genomes Project and activity data from the ENCODE Project. A broad range of transcribed and regulatory nonconserved elements show decreased human diversity, suggesting lineage-specific purifying selection. Conversely, conserved elements lacking activity show increased human diversity, suggesting that some recently became nonfunctional. Regulatory elements under human constraint in nonconserved regions were found near color vision and nerve-growth genes, consistent with purifying selection for recently evolved functions. Our results suggest continued turnover in regulatory regions, with at least an additional 4% of the human genome subject to lineage-specific constraint.
Figures


Comment in
-
Comment on "Evidence of abundant purifying selection in humans for recently acquired regulatory functions".Science. 2013 May 10;340(6133):682. doi: 10.1126/science.1233195. Science. 2013. PMID: 23661742
-
Response to comment on "Evidence of abundant purifying selection in humans for recently acquired regulatory functions".Science. 2013 May 10;340(6133):682. doi: 10.1126/science.1233366. Science. 2013. PMID: 23661743 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

