The molecular profile of luminal B breast cancer
- PMID: 22956860
- PMCID: PMC3430090
- DOI: 10.2147/BTT.S29923
The molecular profile of luminal B breast cancer
Abstract
Molecular profiling studies have found that estrogen receptor-positive (ER+) human breast cancers are comprised of at least two distinct diseases with differing biologies. With the advent of DNA microarrays, global gene expression patterns were used to define the luminal A and luminal B subtypes of ER+ breast cancer, with luminal B cancers showing a more aggressive phenotype including substantially worse outcomes in patients. The luminal B subtype designation could be considered a surrogate for those ER+ tumors having low progesterone receptors, high proliferation, high grade, and predicted poor response to hormone therapy. While they express estrogen receptors, luminal B cancers do not show a corresponding expression of estrogen-regulated genes, and may therefore rely upon alternative pathways for growth. At the molecular level, luminal B cancers appear dramatically distinct from luminal A cancers, at the levels of gene expression, gene copy, somatic mutation, and DNA methylation; luminal B cancers are also genetically and genomically altered to a greater extent than luminal A cancers. While, in the clinical setting, luminal B is typically regarded as an ER+, hormone-sensitive disease, more research is needed into how to better treat it. Comprehensive profiling initiatives, such as The Cancer Genome Atlas, have recently provided us a catalog of mutated or copy altered genes, from which new therapeutic targets could potentially be mined. Candidate pathways that might be targeted in luminal B include those involving growth factor receptors, including HER2 and EGFR, as well as PI3K/Akt/mTor.
Keywords: TCGA; breast cancer; integrative analysis; luminal B; molecular profiling.
Figures




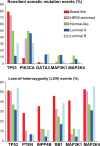

References
-
- Cui X, Schiff R, Arpino G, Osborne CK, Lee AV. Biology of progesterone receptor loss in breast cancer and its implications for endocrine therapy. J Clin Oncol. 2005;23(30):7721–7735. - PubMed
-
- Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

